
Identification of the MEX3 family as potential biomarkers of hepatocellular carcinoma based on bioinformatics and experiments
Highlight box
Key findings
• MEX3 family is associated with the prognosis of hepatocellular carcinoma patients and is related with the tumor microenvironment, including hypoxia and immune responses, which holds significant value for predicting liver cancer outcomes.
What is known and what is new?
• The MEX3 family regulates RNA processing, protein degradation, and oncogenic pathways, is overexpressed in multiple tumors, and exhibits elevated MEX3A expression linked to poor survival outcomes.
• This first comprehensive liver hepatocellular carcinoma (LIHC)-focused study uncovers MEX3 family associations with race-specific expression (MEX3A in Asians), advanced-stage upregulation (MEX3B/C/D), hypoxia-driven mutations, immune infiltration, and prognostic utility (MEX3A as the top biomarker), alongside gene-protein expression discrepancies.
What is the implication, and what should change now?
• MEX3 family has prognostic and diagnostic value for LIHC, guiding personalized treatment.
• The MEX3 family genes have some similarity and redundancy in function, but their roles in tumors vary considerably and the specific roles of each gene and protein need to be investigated.
Introduction
The MEX3 family is a group of RNA binding proteins (RBPs) consisting of four members (MEX3 A/B/C/D) (1). They were initially discovered in the nematode C. elegans and have been found to play a role in embryonic and post-embryonic development (2). Accumulating evidence has highlighted the role of the MEX3 family in various regulatory mechanisms that drive tumorigenesis and cancer progression in such as colorectal cancer (CRC) (3), lung cancer (4), glioma (5), and breast cancer (6). Multiple studies have demonstrated that MEX3 family takes part in cell cycle regulation (7), RNA metabolism (8), post-transcriptional modification (9), hypoxia association (10), immune evasion and tumor metastasis (11). However, the functions of MEX3 RNA binding family members in liver hepatocellular carcinoma (LIHC) remain poorly understood.
The MEX3 family proteins are characterized by two tandem repeat K homology (KH) domains, which are conserved regions that interact with the 3’ untranslated region (UTR) of their target PAL-1 messenger RNA (mRNA). This interaction leads to translational repression of PAL-1, inhibiting its translation (12). In addition to the KH domains, the MEX3 family proteins also contain a ubiquitin E3 ligase RING domain (13). This suggests that E3 ligases not only influence RNA decay but may also have a role in protein fate. The expression of MEX3 family members varies between healthy and malignant tissues of distinct origins, and is associated with disease progression depending on different tumor type (14). Recent studies found that MEX3A, MEX3B, MEX3C and MEX3D have oncogenic roles in a variety of cancers, acting as negative post-translational regulators and degrading tumor suppressor proteins such as RIG-I, RUNX3 and PTEN (9,15-17).
Gene expression includes the transcriptional and post-transcriptional levels. RBPs as highly conserved and abundant proteins function as regulators of alternative splicing, mRNA export, mRNA stability, and translation, which are involved in every aspects of RNA regulation, from transcription to translation (18). RBPs play a crucial role in RNA processes and act as key regulators of cellular homeostasis. The dysregulation of RBPs has been associated with various human diseases, such as cancers (19). In hepatocellular carcinoma, RBPs could contribute to progression via transcription regulation, alternative splicing, alternative polyadenylation, interaction with long non-coding RNAs (lncRNAs), and regulation of mRNA stability (20).
However, only a few studies have investigated the relationship between the MEX3 family and hepatocellular carcinoma (LIHC). Given this research gap, we investigated an extensive collection of databases to explore MEX3 family expression, prognostic value, and immune-related effects in LIHC, thereby offering a more profound understanding of tumor heterogeneity and identify the MEX3 family as a potential molecular marker for prediction. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2095/rc).
Methods
Expression analysis
In this study, we utilized R (version 4.2.1) to analyze the expression levels of the MEX3 family genes in 33 tumor projects. Statistical analysis was performed using the stats (4.2.1) and car (3.1-0) packages, and visualization was done using ggplot2 (3.3.6). RNA sequencing (RNAseq) data from the The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov) was downloaded and organized using the STAR workflow, and the data was extracted in transcripts per million (TPM) format. The Wilcoxon test was used to evaluate the statistical significance of differential expression. To assess the association between the expression levels of MEX3 family genes and clinical variables, TCGA-LIHC samples from the TCGA database were analyzed using R (version 4.2.1). Fisher’s test was used to evaluate the statistical significance of differential expression. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Gene mutation analysis
Gene mutation analysis in MEX3 family genes were examined in LIHC cohorts via cBioPortal (https://www.cbioportal.org/) (21). Associations between mutation profiles and clinicopathological variables were statistically assessed using chi-square tests to determine significance.
Survival analysis
Survival outcomes in LIHC, including overall survival (OS), relapse-free survival (RFS), progression-free survival (PFS), and disease-specific survival (DSS), were stratified by MEX3 family genes expression levels using Kaplan-Meier Plotter (http://kmplot.com/analysis/) (22). The cutoff for low expression versus high expression was determined automatically by the best cutoff model, and arrays with deviation were excluded. The P value (P<0.05) was calculated using the log-rank test.
Correlation and interaction analyses
LinkedOmics (http://www.linkedomics.org/) (23) facilitated visualization of MEX3-associated co-expression networks through volcano plots (800 genes) and heatmaps (top 50 positively/negatively correlated genes). To screen the genes, we initially screened for genes that showed statistically significant correlation differences for each MEX3 family member (P<0.05). Subsequently, we performed a further screening to identify genes with statistically different overall correlations (P<0.05). After that, the genes were sorted based on their correlations, and the top 800 genes were selected for pathway analysis. Spearman’s test was conducted to evaluate the statistical significance. The correlations of MEX3 family gene expression were analyzed using TIMER (http://timer.cistrome.org/) (24).
Gene annotation
GeneCards (https://www.genecards.org/) provided functional annotation for the top 10 positively and negatively correlated motifs linked to each MEX3 family genes’ expression profile.
Functional enrichment analysis
Metascape (https://metascape.org/gp/index.html#/main/step1) (25) mapped enriched GO terms-biological process (BP), cellular component (CC), molecular function (MF)-and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for MEX3-associated co-expressed genes. Complementary analyses in R (version 4.2.1, package clusterProfiler, version 3.14.3) highlighted functional enrichments for the 10 genes exhibiting the strongest interactions with MEX3 family genes.
Immune infiltration analysis
TIMER quantified associations between MEX3 expression and immune cell infiltration, while TISIDB (http://cis.hku.hk/TISIDB/) (26) evaluated links to immune molecule expression in LIHC. Drug sensitivity correlations (chemotherapies/targeted agents) were assessed via GSCA Lite online tool (https://guolab.wchscu.cn/GSCA/#/) (27). Spearman’s tests determined statistical significance for all immune-related analyses.
Survival prediction validation and statistical analysis
Sensitivity and 1−specificity were calculated for each biomarker at different cut-off values. Receiver operating characteristic (ROC) curves were plotted to evaluate the diagnostic performance of the biomarkers. The area under the curve (AUC) was calculated to quantify the discriminatory power of each biomarker. The AUC values of the biomarkers were monitored over time (up to 3 years) to assess the stability of their diagnostic accuracy. A scoring system was developed based on the pathologic T-stage of the disease. Points were assigned to different pathologic T-stages for each biomarker (MEX3A, MEX3B, MEX3C, and MEX3D). The relationship between the total points and the Linear Predictor was established. Based on the scoring system, 1- and 3-year survival probabilities were calculated using appropriate statistical models. A nomogram was developed to predict survival probability based on the biomarker levels and pathologic stage. The observed fraction of survival probability was compared with the nomogram-predicted survival probability for both 1- and 3-year survival to validate the accuracy of the nomogram. All statistical analyses were performed using R (version 4.2.1). The significance level was set at P<0.05.
Real-time quantitative polymerase chain reaction (Q-PCR)
Transformed human liver epithelial-2 (THLE-2) cell was obtained from the FuHeng Cell Center (Shanghai, China), short tandem repeat (STR) profiling was used for authentication. HCC cell lines were obtained from the Chinese Academy of Sciences (Shanghai, China), and cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, USA) with 10% fetal bovine serum (FBS; Gibco BRL), 1% penicillin and 100 µg/mL streptomycin (Gibco BRL) in a humidified incubator containing 5% CO2 at 37 ℃. Cells were harvested for following analyses. RNeasy mini kit (Qiagen, Hilden, Germany) was used to extract cell lines total RNA and using the Quantitate Reverse Transcription Kit (Qiagen) to synthesize complementary DNA (cDNA) according to the manufacturer’s instructions. To quantify MEX3 family genes expression, a FastStart Universal SYBR Green Master kit (Roche Diagnostics, Mannheim, Germany) and a LightCycler 480 (Roche Diagnostics) were used. The primers used in this study were as follows: MEX3A: 5'-TGGAGAACTAGGATGTTTCGGG-3' (F), 5'-GAGGCAGAGTTGATCGAGAGC-3' (R); MEX3B: 5'-GCGGGCGAAGACCAATACTTA-3' (F), 5'-GCCGTGTTCTTATTCCGGGAG-3' (R); MEX3C: 5'-ATGATTCGTGCATCTCGAAACA-3' (F), 5'-GGTCCAACCACTAATCCTACCAC-3' (R); MEX3D: 5'-AGCCGGTCTTCATCGTGAC-3' (F), 5'-GGTCTGTCCGGGAAGGTTG-3' (R); GAPDH: 5'-GTCATCCAACGGGAATGCA-3' (F), 5'-TGATCGGTTACCGTGATCAAAA-3' (R). PCR conditions were as follows: 5 min at 95 ℃, followed by 40 cycles of 95 ℃ for 10 s and 60 ℃ for 60 s. GAPDH was used as an internal control in all PCR reactions. The relative quantities of target gene mRNA compared with an internal control were determined using the 2−ΔΔCt method.
Western blot analysis
The primary antibodies used were as follows: MEX3A (Novus, St. Louis, MI, USA), MEX3B, MEX3C, MEX3D (Santa Cruz, Dallas, TX, USA), and GAPDH (Protein Tech, Wuhan, China). Cells were harvested and lysed in the RIPA buffer (Beyotime, Shanghai, China). The supernatant was collected after centrifugation at 13,000 ×g for 20 min at 4 ℃. The BCA protein kit (Thermo Scientific, Waltham, MA, USA) was used to evaluate protein concentration. All lysates mixed with 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer (Beyotime) were heated for 10 min at 100 ℃; 20 µg of protein samples were separated by SDS-PAGE, and transferred to a polyvinylidene fluoride (PVDF) membrane. First, the PVDF membrane was incubated with QuickBlock Blocking Buffer (Beyotime) for 15 min. Next, the membrane was incubated with primary antibody at 4 ℃ overnight, followed by incubation with horseradish peroxidase-conjugated anti-rabbit/mouse antibody (Beyotime) for 1 hours at room temperature. The protein level was standardized to GAPDH, and then standardized to experimental control. Densitometric analysis of western blot was performed using NIH Image J software (Maryland, USA). All samples derived from the same experiment and that gels/blots were processed in parallel.
Results
Abnormal expression of the MEX3 family genes in pancancer and LIHC patients
We utilized the R (version 4.2.1) to analyze the expression level of the MEX3 family in tumor and paracancerous tissues from TCGA, as well as in normal tissues from genotype-tissue expression (GTEx). The results of the gene expression analysis revealed that MEX3 family genes were differentially expressed in tumors compared to normal or paracancerous tissues across various cancer types. Specifically, MEX3A/B/C/D were significantly upregulated in 17/11/9/19 kinds of tumor tissues, respectively, when compared to 33 paracancerous or normal tissues (Figure 1A-1D). In cholangiocarcinoma (CHOL), head and neck squamous cell carcinoma (HNSC), LIHC, and lung squamous cell carcinoma (LUSC), and stomach adenocarcinoma (STAD) all of MEX3A/B/C/D were statistically significantly increased with P<0.001 (***) (Figure 1A-1D). The OS of MEX3 family members in CHOL, HNSC, and STAD had no statistically significant difference (P>0.05, Figure S1). Therefore, we decided to further investigate the role of the MEX3 family in LIHC.
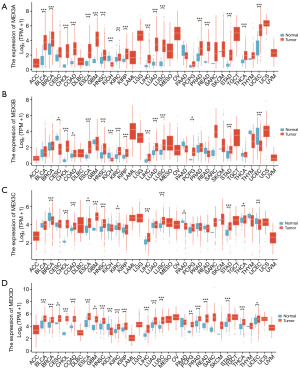
MEX3 family correlation with clinicopathological and prognostic features in LIHC patient
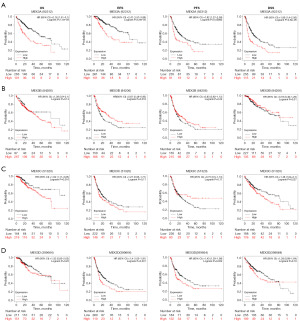
To further explore the association between the expression of MEX3 family and LIHC clinical features, we analyzed the expression levels of MEX3 family with different clinical and pathological features. As shown in Figure 2, MEX3A/B/C/D were high-expressed in LIHC tumor tissues than normal tissues in unpaired and paired groups (Figure 2A,2B). MEX3B/C/D expressed more in higher tumor grade and more advanced clinical stage, while MEX3A showed no significance (Figure 2C,2D). High expression of MEX3 family were all significantly associated with higher alpha fetoprotein (AFP) levels (Figure 2E). Interestingly, we found MEX3A was high-expressed in Asian population than Black or African American and White people, whereas, MEX3B/C/D showed no difference (Figure 2F). As shown in Table 1, high expression of MEX3A was significantly correlated with higher weight (P<0.05), higher height (P<0.05), higher body mass index (BMI) (P<0.01) and higher histologic grade (P<0.01). High expression of MEX3B was significantly associated with patient’s age (P<0.05), gender (P<0.05), and higher histologic grade (P<0.05). High expression of MEX3C was significantly associated with patient’s age (P<0.05), gender (P<0.05), higher pathologic T stage (P<0.05) and tumor status (P<0.05). High expression of MEX3D was significantly associated with higher pathologic T stage (P<0.05), higher weight (P<0.05) and histological type (P<0.05). These results of this study indicated that elevated expression levels of MEX3A might associate with epigenetics phenotypes of patients, and MEX3B/C/D might potentially enhance the aggressiveness of LIHC and contribute to a poorer patients outcomes, further confirming that members of MEX3 family could act as oncogenes in LIHC. To explore this further, we evaluated the correlation between the expression levels of MEX3 family genes and LIHC patient’s prognosis using Kaplan-Meier Plotter. The results showed that high expression of MEX3A gene was significantly associated with worse OS, RFS, PFS and DSS (Figure 3A). High expression of MEX3B gene was significantly associated with better RFS (Figure 3B). High expression of MEX3C gene was significantly associated with worse OS and DSS (Figure 3C). High expression of MEX3D gene was significantly associated with worse RFS and PFS (Figure 3D). These findings suggested that dysregulated expression of the MEX3 family could serve as a potential biomarker for predicting the LIHC prognosis, warranting further experimental validation.
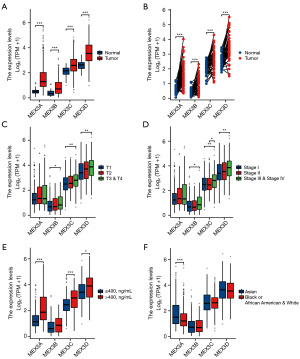
Table 1
| Variables | Expression of MEX3A | Expression of MEX3B | Expression of MEX3C | Expression of MEX3D | Method | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low (n=187) | High (n=187) | P value | Statistic | Low (n=187) | High (n=187) | P value | Statistic | Low (n=187) | High (n=187) | P value | Statistic | Low (n=187) | High (n=187) | P value | Statistic | |||||
| Age (years), n (%) | 0.09 | 2.94 | <0.001 | 15.1 | 0.01 | 6.98 | 0.13 | 2.27 | Chisq test | |||||||||||
| ≤60 | 80 (21.4) | 97 (26.0) | 70 (18.8) | 107 (28.7) | 76 (20.4) | 101 (27.1) | 81 (21.7) | 96 (25.7) | ||||||||||||
| >60 | 106 (28.4) | 90 (24.1) | 117 (31.4) | 79 (21.2) | 111 (29.8) | 85 (22.8) | 105 (28.2) | 91 (24.4) | ||||||||||||
| Gender, n (%) | 0.22 | 1.48 | 0.01 | 7.64 | 0.01 | 7.64 | 0.74 | 0.11 | Chisq test | |||||||||||
| Female | 55 (14.7) | 66 (17.6) | 48 (12.8) | 73 (19.5) | 48 (12.8) | 73 (19.5) | 59 (15.8) | 62 (16.6) | ||||||||||||
| Male | 132 (35.3) | 121 (32.4) | 139 (37.2) | 114 (30.5) | 139 (37.2) | 114 (30.5) | 128 (34.2) | 125 (33.4) | ||||||||||||
| Race, n (%) | 0.001 | 13.26 | 0.67 | 0.81 | 0.52 | 1.29 | 0.76 | 0.56 | Chisq test | |||||||||||
| Asian | 62 (17.1) | 98 (27.1) | 79 (21.8) | 81 (22.4) | 81 (22.4) | 79 (21.8) | 75 (20.7) | 85 (23.5) | ||||||||||||
| Black or African American | 8 (2.2) | 9 (2.5) | 7 (1.9) | 10 (2.8) | 10 (2.8) | 7 (1.9) | 8 (2.2) | 9 (2.5) | ||||||||||||
| White | 108 (29.8) | 77 (21.3) | 96 (26.5) | 89 (24.6) | 86 (23.8) | 99 (27.3) | 94 (26) | 91 (25.1) | ||||||||||||
| Residual tumor, n (%) | 0.22 | – | 0.06 | – | 0.08 | – | 0.90 | – | Fisher test | |||||||||||
| R0 | 168 (48.7) | 159 (46.1) | 172 (49.9) | 155 (44.9) | 169 (49.0) | 158 (45.8) | 165 (47.8) | 162 (47.0) | ||||||||||||
| R1 | 6 (1.7) | 11 (3.2) | 5 (1.4) | 12 (3.5) | 5 (1.4) | 12 (3.5) | 8 (2.3) | 9 (2.6) | ||||||||||||
| R2 | 1 (0.3) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 1 (0.3) | 0 (0.0) | 1 (0.3) | 0 (0.0) | ||||||||||||
| Pathologic stage, n (%) | 0.74 | 1.25 | 0.14 | 5.41 | 0.02 | 10.09 | 0.01 | 11.81 | Yates’ correction | |||||||||||
| Stage I | 92 (26.3) | 81 (23.1) | 93 (26.6) | 80 (22.9) | 97 (27.7) | 76 (21.7) | 102 (29.1) | 71 (20.3) | ||||||||||||
| Stage II | 42 (12.0) | 45 (12.9) | 48 (13.7) | 39 (11.1) | 49 (14.0) | 38 (10.9) | 44 (12.6) | 43 (12.3) | ||||||||||||
| Stage III | 40 (11.4) | 45 (12.9) | 34 (9.7) | 51 (14.6) | 31 (8.9) | 54 (15.4) | 31 (8.9) | 54 (15.4) | ||||||||||||
| Stage IV | 3 (0.9) | 2 (0.6) | 3 (0.9) | 2 (0.6) | 3 (0.9) | 2 (0.6) | 2 (0.6) | 3 (0.9) | ||||||||||||
| Pathologic T stage, n (%) | 0.58 | 1.96 | 0.23 | 4.29 | 0.01 | 10.49 | 0.004 | 13.58 | Chisq test | |||||||||||
| T1 | 98 (26.4) | 85 (22.9) | 98 (26.4) | 85 (22.9) | 102 (27.5) | 81 (21.8) | 107 (28.8) | 76 (20.5) | ||||||||||||
| T2 | 44 (11.9) | 51 (13.7) | 49 (13.2) | 46 (12.4) | 50 (13.5) | 45 (12.1) | 44 (11.9) | 51 (13.7) | ||||||||||||
| T3 | 37 (10.0) | 43 (11.6) | 32 (8.6) | 48 (12.9) | 31 (8.4) | 49 (13.2) | 31 (8.4) | 49 (13.2) | ||||||||||||
| T4 | 7 (1.9) | 6 (1.6) | 7 (1.9) | 6 (1.6) | 3 (0.8) | 10 (2.7) | 3 (0.8) | 10 (2.7) | ||||||||||||
| Pathologic N stage, n (%) | 0.66 | 0.2 | >0.99 | 1.26 | 0.58 | 0.30 | 0.68 | 0.17 | Yates' correction | |||||||||||
| N0 | 124 (48.1) | 130 (50.4) | 126 (48.8) | 128 (49.6) | 131 (50.8) | 123 (47.7) | 122 (47.3) | 132 (51.2) | ||||||||||||
| N1 | 1 (0.4) | 3 (1.2) | 2 (0.8) | 2 (0.8) | 1 (0.4) | 3 (1.2) | 1 (0.4) | 3 (1.2) | ||||||||||||
| Pathologic M stage, n (%) | >0.99 | 1.78 | >0.99 | 4.71 | 1.00 | 4.14 | 0.61 | 0.25 | Yates' correction | |||||||||||
| M0 | 133 (48.9) | 135 (49.6) | 136 (50.0) | 132 (48.5) | 140 (51.5) | 128 (47.1) | 135 (49.6) | 133 (48.9) | ||||||||||||
| M1 | 2 (0.7) | 2 (0.7) | 2 (0.7) | 2 (0.7) | 2 (0.7) | 2 (0.7) | 1 (0.4) | 3 (1.1) | ||||||||||||
| Tumor status, n (%) | 0.14 | 2.16 | 0.74 | 0.11 | 0.04 | 4.34 | 0.06 | 3.60 | Chisq test | |||||||||||
| Tumor free | 107 (30.1) | 95 (26.8) | 102 (28.7) | 100 (28.2) | 111 (31.3) | 91 (25.6) | 109 (30.7) | 93 (26.2) | ||||||||||||
| With tumor | 69 (19.4) | 84 (23.7) | 80 (22.5) | 73 (20.6) | 67 (18.9) | 86 (24.2) | 67 (18.9) | 86 (24.2) | ||||||||||||
| Weight (kg), n (%) | <0.001 | 30.59 | 0.32 | 0.98 | 0.10 | 2.68 | 0.01 | 7.37 | Chisq test | |||||||||||
| ≤70 | 69 (19.9) | 115 (33.2) | 89 (25.7) | 95 (27.5) | 86 (24.9) | 98 (28.3) | 81 (23.4) | 103 (29.8) | ||||||||||||
| >70 | 109 (31.5) | 53 (15.3) | 87 (25.1) | 75 (21.7) | 90 (26.0) | 72 (20.8) | 95 (27.5) | 67 (19.4) | ||||||||||||
| Height (cm), n (%) | <0.001 | 12.66 | 0.66 | 0.19 | 0.34 | 0.91 | 0.51 | 0.43 | Chisq test | |||||||||||
| <170 | 87 (25.5) | 114 (33.4) | 100 (29.3) | 101 (29.6) | 100 (29.3) | 101 (29.6) | 99 (29.0) | 102 (29.9) | ||||||||||||
| ≥170 | 88 (25.8) | 52 (15.2) | 73 (21.4) | 67 (19.6) | 77 (22.6) | 63 (18.5) | 74 (21.7) | 66 (19.4) | ||||||||||||
| BMI (kg/m2), n (%) | <0.001 | 13.55 | 0.77 | 0.09 | 0.24 | 1.38 | 0.05 | 3.70 | Chisq test | |||||||||||
| ≤25 | 74 (22.0) | 103 (30.6) | 89 (26.4) | 88 (26.1) | 86 (25.5) | 91 (27.0) | 81 (24.0) | 96 (28.5) | ||||||||||||
| >25 | 99 (29.4) | 61 (18.1) | 83 (24.6) | 77 (22.8) | 88 (26.1) | 72 (21.4) | 90 (26.7) | 70 (20.8) | ||||||||||||
| Histological type, n (%) | 0.12 | 4.29 | 0.44 | 1.66 | 0.44 | 1.63 | 0.04 | 6.58 | Yates' correction | |||||||||||
| Hepatocellular carcinoma | 182 (48.7) | 182 (48.7) | 184 (49.2) | 180 (48.1) | 183 (48.9) | 181 (48.4) | 183 (48.9) | 181 (48.4) | ||||||||||||
| Hepatocholangiocarcinoma (mixed) | 2 (0.5) | 5 (1.3) | 2 (0.5) | 5 (1.3) | 2 (0.5) | 5 (1.3) | 1 (0.3) | 6 (1.6) | ||||||||||||
| Fibrolamellar carcinoma | 3 (0.8) | 0 (0.0) | 1 (0.3) | 2 (0.5) | 2 (0.5) | 1 (0.3) | 3 (0.8) | 0 (0.0) | ||||||||||||
| Histologic grade, n (%) | <0.001 | 18.26 | 0.02 | 9.52 | 0.19 | 4.72 | 0.21 | 4.48 | Chisq test | |||||||||||
| G1 | 36 (9.8) | 19 (5.1) | 35 (9.5) | 20 (5.4) | 34 (9.2) | 21 (5.7) | 30 (8.1) | 25 (6.8) | ||||||||||||
| G2 | 99 (26.8) | 79 (21.4) | 95 (25.7) | 83 (22.5) | 91 (24.7) | 87 (23.6) | 97 (26.3) | 81 (22.0) | ||||||||||||
| G3 | 49 (13.3) | 75 (20.3) | 50 (13.6) | 74 (20.1) | 55 (14.9) | 69 (18.7) | 53 (14.4) | 71 (19.2) | ||||||||||||
| G4 | 2 (0.5) | 10 (2.7) | 6 (1.6) | 6 (1.6) | 6 (1.6) | 6 (1.6) | 6 (1.6) | 6 (1.6) | ||||||||||||
| AFP (ng/mL), n (%) | <0.001 | 23.56 | 0.04 | 4.08 | <0.001 | 15.39 | 0.03 | 4.64 | Chisq test | |||||||||||
| ≤400 | 130 (46.4) | 85 (30.4) | 120 (42.9) | 95 (33.9) | 129 (46.1) | 86 (30.7) | 122 (43.6) | 93 (33.2) | ||||||||||||
| >400 | 17 (6.1) | 48 (17.1) | 27 (9.6) | 38 (13.6) | 21 (7.5) | 44 (15.7) | 27 (9.6) | 38 (13.6) | ||||||||||||
| Albumin (g/dL), n (%) | 0.14 | 2.23 | 0.27 | 1.19 | 0.37 | 0.81 | 0.11 | 2.62 | Chisq test | |||||||||||
| <3.5 | 42 (14.0) | 27 (9.0) | 41 (13.7) | 28 (9.3) | 34 (11.3) | 35 (11.7) | 42 (14.0) | 27 (9.0) | ||||||||||||
| ≥3.5 | 117 (39.0) | 114 (38.0) | 120 (40.0) | 111 (37.0) | 128 (42.7) | 103 (34.3) | 115 (38.3) | 116 (38.7) | ||||||||||||
| Prothrombin time, n (%) | 0.25 | 1.33 | 0.39 | 0.73 | 0.66 | 0.20 | 0.52 | 0.42 | Chisq test | |||||||||||
| ≤4 | 104 (35.0) | 104 (35.0) | 108 (36.4) | 100 (33.7) | 111 (37.4) | 97 (32.7) | 106 (35.7) | 102 (34.3) | ||||||||||||
| >4 | 51 (17.2) | 38 (12.8) | 51 (17.2) | 38 (12.8) | 45 (15.2) | 44 (14.8) | 49 (16.5) | 40 (13.5) | ||||||||||||
| Child-Pugh grade, n (%) | 0.64 | – | 0.56 | – | 0.41 | – | 0.42 | – | Fisher test | |||||||||||
| A | 122 (50.6) | 97 (40.2) | 126 (52.3) | 93 (38.6) | 132 (54.8) | 87 (36.1) | 122 (50.6) | 97 (40.2) | ||||||||||||
| B | 10 (4.1) | 11 (4.6) | 13 (5.4) | 8 (3.3) | 11 (4.6) | 10 (4.1) | 10 (4.1) | 11 (4.6) | ||||||||||||
| C | 1 (0.4) | 0 (0.0) | 0 (0.0) | 1 (0.4) | 0 (0.0) | 1 (0.4) | 0 (0.0) | 1 (0.4) | ||||||||||||
| Fibrosis Ishak score, n (%) | 0.93 | 0.84 | 0.62 | 2.64 | 0.72 | 2.08 | 0.32 | 4.69 | Yates' correction | |||||||||||
| 0 | 43 (20.0) | 32 (14.9) | 39 (18.1) | 36 (16.7) | 47 (21.9) | 28 (13.0) | 40 (18.6) | 35 (16.3) | ||||||||||||
| 1/2 | 16 (7.4) | 15 (7.0) | 17 (7.9) | 14 (6.5) | 16 (7.4) | 15 (7.0) | 15 (7.0) | 16 (7.4) | ||||||||||||
| 3/4 | 16 (7.4) | 12 (5.6) | 11 (5.1) | 17 (7.9) | 16 (7.4) | 12 (5.6) | 13 (6.0) | 15 (7.0) | ||||||||||||
| 5 | 5 (2.3) | 4 (1.9) | 5 (2.3) | 4 (1.9) | 4 (1.9) | 5 (2.3) | 3 (1.4) | 6 (2.8) | ||||||||||||
| 6 | 44 (20.5) | 28 (13.0) | 41 (19.1) | 31 (14.4) | 44 (20.5) | 28 (13.0) | 45 (20.9) | 27 (12.6) | ||||||||||||
| Vascular invasion, n (%) | 0.41 | 0.68 | 0.53 | 0.40 | 0.61 | 0.26 | 0.13 | 2.30 | Chisq test | |||||||||||
| No | 116 (36.5) | 92 (28.9) | 108 (34.0) | 100 (31.4) | 114 (35.8) | 94 (29.6) | 115 (36.2) | 93 (29.2) | ||||||||||||
| Yes | 56 (17.6) | 54 (17.0) | 53 (16.7) | 57 (17.9) | 57 (17.9) | 53 (16.7) | 51 (16.0) | 59 (18.6) | ||||||||||||
| Adjacent hepatic tissue inflammation, n (%) | 0.25 | 2.79 | 0.70 | 0.70 | 0.76 | 0.56 | 0.95 | 0.11 | Chisq test | |||||||||||
| None | 69 (29.1) | 49 (20.7) | 58 (24.5) | 60 (25.3) | 67 (28.3) | 51 (21.5) | 61 (25.7) | 57 (24.1) | ||||||||||||
| Mild | 50 (21.1) | 51 (21.5) | 54 (22.8) | 47 (19.8) | 53 (22.4) | 48 (20.3) | 52 (21.9) | 49 (20.7) | ||||||||||||
| Severe | 12 (5.1) | 6 (2.5) | 8 (3.4) | 10 (4.2) | 9 (3.8) | 9 (3.8) | 10 (4.2) | 8 (3.4) | ||||||||||||
If the data contains infinite values, the infinite values are treated as missing. Missing variables are not handled uniformly. AFP, alpha fetoprotein; BMI, body mass index; LIHC, liver hepatocellular carcinoma.
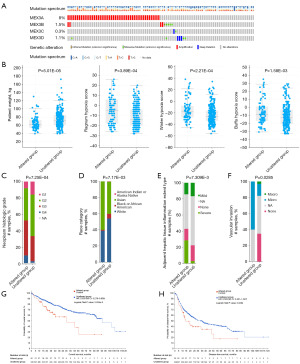
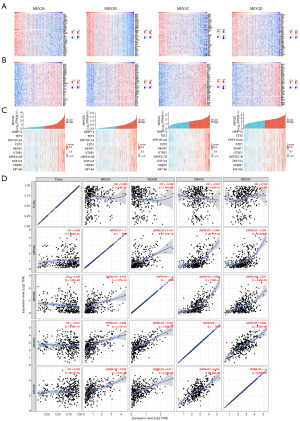
MEX3 family gene mutations and their association with clinicopathological features in LIHC patients
To investigate the mechanism underlying the differential expression of the MEX3 family genes in LIHC, we utilized the cBioPortal online tool to analyze mutations in the MEX3 family genes. Among LIHC patients, 62 samples (10.33%) exhibited two or more types of genetic alterations in the MEX3 family genes (Figure 4A). The most frequent alteration was amplification in the MEX3A gene (8%). In addition, MEX3B and MEX3D showed mutation rates of 1.5% and 1.1%, respectively, and the most common type of variant was amplification, missense mutations and deep deletion, while only 1 missense mutation and 1 deep deletion mutation were detected in the MEX3C gene. We further compared the clinicopathological features of LIHC patients with and without MEX3 family gene mutations. The analysis revealed that patients with a lower body weight, hypoxia condition, higher neoplasms histological grade and Asian population were more prone to MEX3 family gene mutations (Figure 4B-4D). Additionally, patients with MEX3 family gene mutations exhibited a more serves inflammation in adjacent hepatic tissues (Figure 4E) and a higher incidence of micro vascular invasion (Figure 4F). Prognostic analysis indicated that MEX3 family gene mutations significantly impacted OS (Figure 4G) and disease-free survival (DFS; Figure 4H) in LIHC patients. In summary, MEX3 family gene mutations were more prevalent in LIHC patients with lower body weight and hypoxia conditions. These mutations were associated with more aggressive tumor behavior, higher histological grades, and poorer patient outcomes, suggesting their potential role in driving malignancy and influencing prognosis in LIHC.
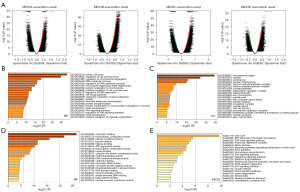
Functional enrichment analysis of the MEX3 family and co-expressed genes in LIHC patients
To investigate the functional interaction between MEX3 family genes and their co-expressed genes in LIHC, we identified 800 co-expressed genes associated with the MEX3 family using the LinkedOmics database. The differential expression patterns of these genes were visualized in a volcano plot (Figure 5A). Subsequently, we performed Gene Ontology (GO) and KEGG pathway analyses on these 800 genes to elucidate the potential biological roles and mechanisms of the MEX3 family in the initiation and progression of LIHC. The GO analysis results showed significant enrichment of the BP terms “mitotic cell cycle”, “regulation of cell cycle process” and “regulation of chromosome segregation” (Figure 5B). The enriched CC terms mainly included “chromosomal region”, “spindle” and “centrosome” (Figure 5C). The enriched MF terms mainly included “chromatin binding”, “transcription coregulator activity” and “histone modifying activity” (Figure 5D). The KEGG pathway analysis showed that the target genes were mainly associated with the terms “Cell cycle”, “ATP-dependent chromatin remodeling”, and “TGF-beta signaling pathway” (Figure 5E). To further investigate the functional mechanisms of the MEX3 family and its co-expressed genes, we generated heatmaps to visualize the top 50 positive-correlated genes (Figure 6A) and the top 50 negative-correlated genes (Figure 6B) with MEX3 family genes. Functional annotation of the positive-correlated genes revealed their involvement in transcriptional regulation (TCF3, FBXO46, ZBTB12, ARHGEF2, LZTS2, ZNF74, GEM, ZNF532, CBL, CREB1, NCOA6, PAXIP1, DOT1L), protein modification (MIB1, CBL, RNF138, NEDD1), epitope modification (PYGO2, ESCO1, SMCHD1, RASSF3, DVL3, TNRC18, DOT1L), cell cycle regulation (MARVELD1, IQGAP1, CDK2), apoptosis (FAM189B), cell adhesion (PCDH18), and immune response (COL3A1, ADAM17). Meanwhile, the functions of negatively related genes included metabolism regulation (LDHD, MLYCD, AASS, AGXT2L1, CTH, COQ9, MRPL40, DCXR, DHRS4, TACO1, CISD3, BLVRB, DCI, UQCRQ, ETFB, MRPS5, OXSM), protein modification (GABARAPL1, UQCRC1, SOD1, ISOC2, ZNHIT1), and autophagy (PRKAG2). Notably, our analysis showed that the MMP14, TET3, PPP1R12A, FZD1, SENP1, GTSE1, DEPDC1B, ZNF124, CREB1, KIF18A were positively related to the expression of MEX3 family genes (Figure 6C). Subsequently, we utilized the TIMER database to investigate the relationships among MEX3 family members. The results indicated that the expression levels of each member were significantly positively correlated with one another, with the strongest correlation observed between MEX3B and MEX3C (cor =0.638) (Figure 6D). Based on these results, we hypothesized that the MEX3 family genes may collaborate with diverse transcriptional regulators, protein-modifying factors, metabolism, and immune modification to participate in the occurrence and development of LIHC.
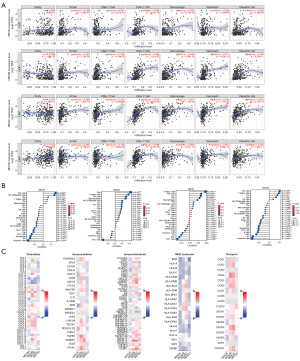
Association of the MEX3 family with tumor-infiltrating immune cells and immune molecules in LIHC patients
Consistent with above findings, we hypothesized that the MEX3 family could influence the tumor microenvironment in LIHC patients by regulating immune cells and immune molecules. For further investigation, we utilized the TIMER database to examine the correlation between the expression of MEX3 family genes and the infiltration level of various lymphocytes. The analysis revealed that elevated expression of MEX3 family genes was associated with increased infiltration of B cells, CD8+ T cells (with the exception of MEX3A), CD4+ T cells, macrophages, neutrophils, dendritic cells and other types of infiltrating lymphocytes (Figure 7A). In the meantime, we used the TCGA data to look more closely at the relationship between MEX3 family gene expression and lymphocyte infiltration levels. The results showed that MEX3A gene expression was associated with increased Th2 cells infiltration, MEX3B was Tem cells, MEX3C was T helper cells, and MEX3D was Th2 cells (Figure 7B). Whereas, MEX3A was negatively associated with cytotoxic cells and dendritic cells (DC), MEX3B was Th17 cells and DC, MEX3C was Th17 cells and plasmacytoid dendritic cell (pDC), MEX3D was cytotoxic cells and DC. Next, we analyzed the correlation between MEX3 family expression and chemokines, immunoinhibitor, immunostimulator, major histocompatibility complex (MHC) molecular, receptor (Figure 7C) in LIHC using the TISIDB database. The results showed that MEX3 family genes were positively associated with an increase in the expression of immunostimulatory molecules such as CD276, HHLA2, TNFRSF9, TNFRSF15, TNFRSF4 and ULBP1, immunosuppressive molecules such as VTCN1, and chemokines such as CCL28. Besides, MEX3B and MEX3C was positively associated with more molecules, such as MHC molecules HLA-DOA, HLA-DQ1 and HLA-DQ2, chemokine receptors CCR1, CCR2 and CXCR4, etc. These provides important information for exploring the role of MEX3 family in tumor immune microenvironment and immunotherapy.
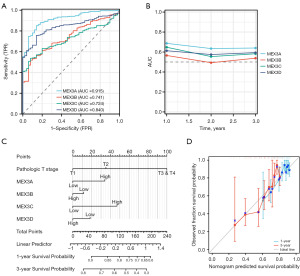
The value of the MEX3 family in LIHC prognostic diagnosis
We further investigated the value of the MEX3 Family in LIHC prognostic diagnosis. ROC analysis was used to evaluate the prognostic evaluation ability of the MEX3 family genes in LIHC, and the AUC values of MEX3A/B/C/D were 0.915, 0.741, 0.724, and 0.840, respectively (Figure 8A). The time-dependent ROC curves showed that MEX3A had the highest AUC value at all time points, indicating that it consistently showed better predictive power over the 3 years (Figure 8B). The AUC value for MEX3B is relatively low throughout the time period, but picks up in the third year (Figure 8B). The AUC values for MEX3C and MEX3D are relatively similar, remaining stable throughout the time period with no significant downward or upward trend (Figure 8B). Nomogram figure shows 1- and 3-year survival probabilities of MEX3 family members at different pathological T stages for total points and linear predictor variables (Figure 8C). Expression levels of MEX3 family members in relation to pathological stage and their relationship impacts patient survival probability. Elevated expression of MEX3 family members was associated with higher pathological stage and lower survival probability. Calibration plots showed that the 3-year OS probability predicted by the nomogram was closer to the ideal OS probability than 1-year OS probability (Figure 8D). Thus, the MEX3 family might represent novel biomarkers for predicting LIHC prognosis.
Protein expression levels of MEX3 family in LIHC
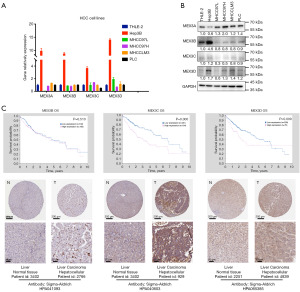
We investigated the mRNA and protein levels of MEX3 family members in LIHC cell lines. The Q-PCR showed that MEX3 family member mRNAs were all highly expressed in Hep3B cells, with insignificant differences in expression levels in other LIHC cells, especially when compared to normal hepatocyte THLE-2 (Figure 9A). Western blot showed that MEX3A protein was highly expressed in LIHC cell lines, MEX3B was significantly higher in Hep3B cells, and the difference between MEX3C and MEX3D was not significant (Figure 9B). Moreover, protein expression data from the Human Protein Atlas project showed that high expression of MEX3C/D protein was significantly associated with worse OS (Figure 9C), and immunohistochemistry (IHC) results of LIHC patients showed MEX3C/D proteins were obviously higher expressed in tumor tissues. These results indicated that variability in mRNA and protein expression levels of MEX3 family members in LIHC.
Discussion
The MEX3 family, constituting evolutionarily-conserved RNA-binding proteins with additional ubiquitin E3 ligase properties, mediates post-transcriptional regulation across various organisms (28). In recent years, an increased understanding of the oncogenic role of the MEX3 family has been gained, and related studies have unveiled a potential cancer-promoting role in a variety of malignant tumors, including cervical cancer, lung cancer, CRC (29-31). However, the elucidation of the role and mechanism of the MEX3 family in hepatocellular carcinoma pathogenesis remains scant, and detailed bioinformatics studies have not been conducted. This study pioneers an in-depth analysis of the expression, gene mutation, association with immune cell infiltration, and prognostic significance of the MEX3 family in LIHC. By utilizing several public databases, we uncovered that the expression levels of MEX3A, MEX3B, MEX3C and MEX3D-all members of the MEX3 family-were significantly elevated in LIHC. Additionally, the expression of MEX3 family members was closely tied to tumor stage, tumor grade, treatment response, and other clinicopathological factors. These findings indicate that MEX3 family members have substantial potential as prognostic and diagnostic biomarkers for LIHC patients. Furthermore, we unveiled a potential mechanism through which the MEX3 family contributes to the pathogenesis and progression of LIHC and its interaction with the tumor immune response, thereby offering potential therapeutic targets for LIHC and bearing significant clinical implications.
Research has shown that MEX3A influenced G1 phase cell cycle by regulating CDK2 expression in CRC (32). Similarly, our study revealed that the expression of MEX3A associated with cell cycle molecules CDK1, and the GO and KEGG analysis showed their co-expressed genes significant enriched in the BP terms “mitotic cell cycle”, and the KEGG pathway “Cell cycle” in LIHC. Recent study found that hsa-miR-6887-3p binding to the MEX3A mRNA to suppress MEX3A expression which in turn downregulated the interaction between MEX3A and RAP1 GTPase activating protein (RAP1GAP) of MEK/ERK/HIF-1α/MAPK pathway in CRC (33). MAPK pathway could active CDK2 and regulate G1/S transition and S phase progression resulting in breast cancer cell proliferation (34). Likewise, MEX3A could promote migration and proliferation of breast cancer cells via activating RhoA/ROCK1/LIMK1 signaling pathway which contributes to increased expression of c-myc avian myelocytomatosis viral oncogene homolog (c-MYC) and cyclin D1, leading to cell proliferation and migration (35,36). Moreover, MEX3A could regulate nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), β-catenin and Ak strain transforming (AKT) signaling pathway to manipulate cancer cell proliferation, epithelial to mesenchymal transition, invasion, migration, and apoptosis (6,37-39). In addition, MEX3A could facilitate CRC angiogenesis by activating the glycolytic pathway (31). Our GO and KEGG analyses of the MEX3A family and its 800 co-expressed genes in LIHC suggest that they play a role in many BPs, such as “mitotic cell cycle”, “ECM-receptor interaction”, “Wnt signaling pathway”, “growth”, and “positive regulation of cellular component biogenesis” which is consistent with the finding that overexpression of MEX3A promotes the proliferation, invasion, metastasis, and metabolism of many kinds of tumors, suggesting MEX3A as a potential treatment target for cancer therapy. As a RING-type E3 ubiquitin ligase, MEX3A facilitates the ubiquitination and degradation of RIG-I, an essential innate immune receptor, to inhibit IRF3-mediated innate antiviral immune response (40,41). Interestingly, MEX3A can lead to tumor cell chemotherapy resistance of temozolomide in glioblastoma by impairing DNA mismatch repair signaling (8). These suggest MEX3A likely plays a key role in regulating the immune response of hepatocellular carcinoma cells and inducing chemotherapy resistance.
MEX3B regularly functioned as an E3 ubiquitin ligase, which involved in the ubiquitination of a tumor suppressor, runt-related transcription factor 3 (RUNX3), leading to its degradation in gastric cancers (42). MEX3B also associated with a poor prognosis of hepatitis B virus (HBV)-associated hepatocellular carcinoma for being an important cellular target for the design of epigenetic therapies for chronic HBV infection (43). Meanwhile, the immunosuppressive cytokine IL-37b inhibits MEX3B, thereby suppressing TLR3, a key component of innate and adaptive immunity, and thus reducing inflammation in chronic rhinosinusitis patients (44). Recently, MEX3B was demonstrated to potentiate the K63-linked ubiquitination of TLR3, and then elicits the IRF3-mediated antiviral innate immune responses in grass carp (28). In addition, MEX3B could modulate stress-induced apoptosis through post-transcriptional gene regulation of the pro-apoptotic BIM gene, which is induced by MYC and leads to apoptosis by activation of BAX/BAK (45,46). Besides, MEX3B also regulates the localization of the RAP1GAP to play a crucial role in the RAP1 pathway, which is essential for normal Sertoli cell physiology and the formation of the blood-testis barrier (47). MEX3B regulates the dynamic exchange of mRNPs between processing bodies and stress granules through its interaction with 14-3-3 adaptors, and plays a key role in the regulation of RNA granules (48). MEX3B reduces TGFBR3 mRNA stability by preferentially binding to the 3’ UTR, leading to downregulation of TGF-β R3 expression in nasal epithelial cells (49). These studies have revealed that MEX3B regulates the tumor progress and immune response via protein modulation and post-transcriptional regulation, and MEX3B may be a marker of malignant tumor progression and a potential therapeutic target.
Lately, the study of MEX3C in hepatocellular carcinoma found that MEX3C triggered the degradation of SOCS3 mRNA, which in turn stimulated the activation of JAK2/STAT3 and tumor metastasis (50). Knockdown of MEX3C combined with lenvatinib showed a stronger inhibitory effect, suggesting that MEX3C could be a potential target for the treatment of hepatocellular carcinoma (51). Additionally, MEX3C could catalyze K27-linked polyubiquitination of tumor suppressor PTEN and promote EMT of diabetic kidney disease (52). In bladder cancer, MEX3C promote tumorigenesis by regulating lipid metabolism through MAPK/JNK pathway (53). The mutation of MEX3C led to leanness, improved blood glucose and lipid profiles in transgenic mice, and increased energy expenditure independent of physical activity (54). Furthermore, MEX3C also regulated posttranscription and RNA degradation (55). MEX3C could bind to the 3’-UTR of the human leukocyte antigen serotype (HLA-A2) mRNA and mediate MHC-I mRNA degradation (13,56,57). This study provides additional strong evidence for the role of MEX3C in immune modulation. Therefore, we speculate that MEX3C may be an oncogene in hepatocellular carcinoma through mRNA and protein degradation, and be expected to a novel therapeutic target for hepatocellular carcinoma.
MEX3D was found to exhibit oncogenic properties in a variety of cancer types. Using a replication-incompetent lentiviral vector for an insertional mutagenesis screen in the progression to androgen-independent prostate cancer, the study identified MEX3D that influenced the disease (58). MEX3D destabilized TSC22D1 mRNA levels and promoted cell proliferation and inhibited apoptosis in cervical cancer development, suggesting that MEX3D could be a potential therapeutic target for cervical cancer (29). Additionally, loss of PTEN strongly upregulates expression of MEX3D and its target TCF3 and promotes transformation associated phenotypes via this pathway in prostate cancer (17). Nevertheless, fewer studies have been conducted on the role of MEX3D in cancer progression. Our studies showed MEX3D was related to the expression of CDK2 and its protein expression level was upregulated in LIHC, suggesting that MEX3D might play a role in the regulation of cell cycle, and more related studies should be performed.
Since first discovery of MEX3 family, their function, such as post-transcriptional regulation, stem cell self-renewal and differentiation and post-transcriptional regulation, were gradually revealed. However, the related researches of MEX3 family were limited and there was rare evidence to demonstrate the relationship of MEX3 family and tumor progress. Therefore, we speculated the undermined mechanisms between MEX3 family and LIHC. The MEX3 family has been found implicated in hepatocellular carcinoma progression. MEX3A and B have been shown to promote tumorigenesis and cell proliferation, suggesting their roles as potential oncogenes. MEX3C, associated with tumor metastasis, could serve as a therapeutic target. MEX3D’s upregulation in LIHC indicates its potential role in cell cycle regulation. Future research should focus on understanding the molecular mechanisms underlying these effects and validating the potential of MEX3 proteins as therapeutic targets. Their clinical significance lies in the potential to advance personalized treatment strategies for hepatocellular carcinoma.
Conclusions
In the results of our bioinformatics analyses, MEX3 family showed a close relation to the onset, progression and prognosis of hepatocellular carcinoma which indicated MEX3 family could be prognostic markers and new therapeutic targets for hepatocellular carcinoma patients. There is an urgent need for more foundational research studies on MEX3 family in hepatocellular carcinoma, both in vivo and in vitro.
Acknowledgments
We are very grateful to the Prof Lili Cai from Cancer Institute, Longhua Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, who supported the data analysis.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2095/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2095/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2095/prf
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2095/coif). J.X. receives support funding from the National Natural Science Foundation of China (No. 82402682), Natural Science Foundation of Shanghai (No. 23ZR1411100), Youth Fund of Zhongshan Hospital Affiliated Fudan University (No. 2022ZSQN12); W.S. receives support funding from the National Key Research and Development Program of China (No. 2022YFC3602301); B.W. receives support funding from the National Key Research and Development Program of China (No. 2022YFC3602302), Specialized Fund for the Clinical Researches of Zhongshan Hospital Affiliated to Fudan University (No. 2020ZSLC54), the Projects from Excellent Backbone of Zhongshan Hospital, Fudan University (No. 2021ZSGG08); W.G. receives support funding from the National Natural Science Foundation of China (Nos. 81972000, 81902139), the Constructing Project of Clinical Key Disciplines in Shanghai (No. shslczdzk03302), Shanghai Baoshan District Medical Key Specialty Project (No. BSZK-2023-A18), Key Medical and Health Projects of Xiamen (No. YDZX20193502000002), Shanghai Municipal Science and Technology Commission Innovation Action Chemical Reagent Development Project (No. 21142201600), Shanghai Professional and Technical Service Platform Construction (No. 22DZ2291900); W.Y. receives support funding from the National Natural Science Foundation of China (No. 82172348). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang NN, Mootz DE, Walhout AJ, et al. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development 2002;129:747-59. [Crossref] [PubMed]
- Draper BW, Mello CC, Bowerman B, et al. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 1996;87:205-16. [Crossref] [PubMed]
- Chen RX, Xu SD, Deng MH, et al. Mex-3 RNA binding family member A (MEX3A)/circMPP6 complex promotes colorectal cancer progression by inhibiting autophagy. Signal Transduct Target Ther 2024;9:80. [Crossref] [PubMed]
- Liang J, Li H, Han J, et al. Mex3a interacts with LAMA2 to promote lung adenocarcinoma metastasis via PI3K/AKT pathway. Cell Death Dis 2020;11:614. [Crossref] [PubMed]
- Yang C, Zhan H, Zhao Y, et al. MEX3A contributes to development and progression of glioma through regulating cell proliferation and cell migration and targeting CCL2. Cell Death Dis 2021;12:14. [Crossref] [PubMed]
- Wang Y, Liang Q, Lei K, et al. Targeting MEX3A attenuates metastasis of breast cancer via β-catenin signaling pathway inhibition. Cancer Lett 2021; Epub ahead of print. [Crossref]
- Qiu Y, Meng M, Cao C, et al. RNA-binding protein MEX3A controls G1/S transition via regulating the RB/E2F pathway in clear cell renal cell carcinoma. Mol Ther Nucleic Acids 2022;27:241-55. [Crossref] [PubMed]
- Gan T, Wang Y, Xie M, et al. MEX3A Impairs DNA Mismatch Repair Signaling and Mediates Acquired Temozolomide Resistance in Glioblastoma. Cancer Res 2022;82:4234-46. [Crossref] [PubMed]
- He Z, Zhang H, Xiao H, et al. Ubiquitylation of RUNX3 by RNA-binding ubiquitin ligase MEX3C promotes tumorigenesis in lung adenocarcinoma. J Transl Med 2024;22:216. [Crossref] [PubMed]
- Hu B, Yang XB, Sang XT. Development and Verification of the Hypoxia-Related and Immune-Associated Prognosis Signature for Hepatocellular Carcinoma. J Hepatocell Carcinoma 2020;7:315-30. [Crossref] [PubMed]
- Yang K, Chen G, Yu F, et al. Molecular mechanism of specific HLA-A mRNA recognition by the RNA-binding-protein hMEX3B to promote tumor immune escape. Commun Biol 2024;7:158. [Crossref] [PubMed]
- Zhu SJ, Hallows SE, Currie KW, et al. A mex3 homolog is required for differentiation during planarian stem cell lineage development. Elife 2015;4:e07025. [Crossref] [PubMed]
- Yang L, Wang C, Li F, et al. The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity. J Biol Chem 2017;292:16221-34. [Crossref] [PubMed]
- Buchet-Poyau K, Courchet J, Le Hir H, et al. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res 2007;35:1289-300. [Crossref] [PubMed]
- Bufalieri F, Caimano M, Lospinoso Severini L, et al. The RNA-Binding Ubiquitin Ligase MEX3A Affects Glioblastoma Tumorigenesis by Inducing Ubiquitylation and Degradation of RIG-I. Cancers (Basel) 2020;12:321. [Crossref] [PubMed]
- Huang L, Malu S, McKenzie JA, et al. The RNA-binding Protein MEX3B Mediates Resistance to Cancer Immunotherapy by Downregulating HLA-A Expression. Clin Cancer Res 2018;24:3366-76. [Crossref] [PubMed]
- Shao L, Wang J, Karatas O, et al. MEX3D is an oncogenic driver in prostate cancer. Prostate 2021;81:1202-13. [Crossref] [PubMed]
- Li W, Deng X, Chen J. RNA-binding proteins in regulating mRNA stability and translation: roles and mechanisms in cancer. Semin Cancer Biol 2022;86:664-77. [Crossref] [PubMed]
- Yao ZT, Yang YM, Sun MM, et al. New insights into the interplay between long non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun (Lond) 2022;42:117-40. [Crossref] [PubMed]
- Zhang K, Barry AE, Lamm R, et al. The role of RNA binding proteins in hepatocellular carcinoma. Adv Drug Deliv Rev 2022;182:114114. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Győrffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation (Camb) 2024;5:100625. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics 2018;34:3771-2. [Crossref] [PubMed]
- Hu J, Xiang Y, Zhu X, et al. Grass carp (Ctenopharyngodon idella) Mex3B positively regulates innate immunity by promoting the K63-linked ubiquitination of TLR3. Fish Shellfish Immunol 2023;141:109023. [Crossref] [PubMed]
- Zheng Z, Chen X, Cai X, et al. RNA-binding protein MEX3D promotes cervical carcinoma tumorigenesis by destabilizing TSC22D1 mRNA. Cell Death Discov 2022;8:250. [Crossref] [PubMed]
- Zhang M, Cao L, Hou G, et al. Investigation of the Potential Correlation Between RNA-Binding Proteins in the Evolutionarily Conserved MEX3 Family and Non-small-Cell Lung Cancer. Mol Biotechnol 2023;65:1263-74. [Crossref] [PubMed]
- Lu Y, Bi T, Zhou S, et al. MEX3A promotes angiogenesis in colorectal cancer via glycolysis. Libyan J Med 2023;18:2202446. [Crossref] [PubMed]
- Zhou X, Li S, Ma T, et al. MEX3A knockdown inhibits the tumorigenesis of colorectal cancer via modulating CDK2 expression. Exp Ther Med 2021;22:1343. [Crossref] [PubMed]
- Li H, Liang J, Wang J, et al. Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p. Cancer Commun (Lond) 2021;41:472-91. [Crossref] [PubMed]
- Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res 2002;12:9-18. [Crossref] [PubMed]
- Yan L, Li H, An W, et al. Mex-3 RNA binding MEX3A promotes the proliferation and migration of breast cancer cells via regulating RhoA/ROCK1/LIMK1 signaling pathway. Bioengineered 2021;12:5850-8. [Crossref] [PubMed]
- Wang T, Rao D, Yu C, et al. RHO GTPase family in hepatocellular carcinoma. Exp Hematol Oncol 2022;11:91. [Crossref] [PubMed]
- Jiang H, Zhang X, Luo J, et al. Knockdown of hMex-3A by small RNA interference suppresses cell proliferation and migration in human gastric cancer cells. Mol Med Rep 2012;6:575-80. [Crossref] [PubMed]
- Xiang XX, Liu YL, Kang YF, et al. MEX3A promotes nasopharyngeal carcinoma progression via the miR-3163/SCIN axis by regulating NF-κB signaling pathway. Cell Death Dis 2022;13:420. [Crossref] [PubMed]
- Xu Y, Pan S, Chen H, et al. MEX3A suppresses proliferation and EMT via inhibiting Akt signaling pathway in cervical cancer. Am J Cancer Res 2021;11:1446-62.
- Clues to Estrogen and Breast Cancer Cell Survival Involve NEDD8. SENP8, SirT1, NFκB and a Breast Cancer Associated Protein BCA3. Cancer Biology & Therapy 2006;5:1262.
- Jiang Z, Sun Z, Hu J, et al. Grass Carp Mex3A Promotes Ubiquitination and Degradation of RIG-I to Inhibit Innate Immune Response. Front Immunol 2022;13:909315. [Crossref] [PubMed]
- Xue M, Chen LY, Wang WJ, et al. HOTAIR induces the ubiquitination of Runx3 by interacting with Mex3b and enhances the invasion of gastric cancer cells. Gastric Cancer 2018;21:756-64. [Crossref] [PubMed]
- Zhang H, Xing Z, Mani SK, et al. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology 2016;64:1033-48. [Crossref] [PubMed]
- Liu JX, Liao B, Yu QH, et al. The IL-37-Mex3B-Toll-like receptor 3 axis in epithelial cells in patients with eosinophilic chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol 2020;145:160-72. [Crossref] [PubMed]
- Oda T, Yamazumi Y, Hiroko T, et al. Mex-3B induces apoptosis by inhibiting miR-92a access to the Bim-3'UTR. Oncogene 2018;37:5233-47. [Crossref] [PubMed]
- Muthalagu N, Junttila MR, Wiese KE, et al. BIM is the primary mediator of MYC-induced apoptosis in multiple solid tissues. Cell Rep 2014;8:1347-53. [Crossref] [PubMed]
- Le Borgne M, Chartier N, Buchet-Poyau K, et al. The RNA-binding protein Mex3b regulates the spatial organization of the Rap1 pathway. Development 2014;141:2096-107. [Crossref] [PubMed]
- Courchet J, Buchet-Poyau K, Potemski A, et al. Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J Biol Chem 2008;283:32131-42. [Crossref] [PubMed]
- Liu JX, Chen AN, Yu Q, et al. MEX3B inhibits collagen production in eosinophilic nasal polyps by downregulating epithelial cell TGFBR3 mRNA stability. JCI Insight 2023;8:e159058. [Crossref] [PubMed]
- Xiao Y, Li Y, Shi D, et al. MEX3C-Mediated Decay of SOCS3 mRNA Promotes JAK2/STAT3 Signaling to Facilitate Metastasis in Hepatocellular Carcinoma. Cancer Res 2022;82:4191-205. [Crossref] [PubMed]
- Guo J, Zhao J, Xu Q, et al. MEX3C as a potential target for hepatocellular carcinoma drug and immunity: combined therapy with Lenvatinib. BMC Cancer 2023;23:967. [Crossref] [PubMed]
- Li Y, Hu Q, Li C, et al. PTEN-induced partial epithelial-mesenchymal transition drives diabetic kidney disease. J Clin Invest 2019;129:1129-51. [Crossref] [PubMed]
- Chao H, Deng L, Xu F, et al. MEX3C regulates lipid metabolism to promote bladder tumorigenesis through JNK pathway. Onco Targets Ther 2019;12:3285-94. [Crossref] [PubMed]
- Jiao Y, George SK, Zhao Q, et al. Mex3c mutation reduces adiposity and increases energy expenditure. Mol Cell Biol 2012;32:4350-62. [Crossref] [PubMed]
- Jiao Y, Bishop CE, Lu B. Mex3c regulates insulin-like growth factor 1 (IGF1) expression and promotes postnatal growth. Mol Biol Cell 2012;23:1404-13. [Crossref] [PubMed]
- Cano F, Rapiteanu R, Sebastiaan Winkler G, et al. A non-proteolytic role for ubiquitin in deadenylation of MHC-I mRNA by the RNA-binding E3-ligase MEX-3C. Nat Commun 2015;6:8670. [Crossref] [PubMed]
- Cano F, Bye H, Duncan LM, et al. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J 2012;31:3596-606. [Crossref] [PubMed]
- Schinke EN, Bii V, Nalla A, et al. A novel approach to identify driver genes involved in androgen-independent prostate cancer. Mol Cancer 2014;13:120. [Crossref] [PubMed]


