
Identification of a novel prognostic gene signature in pleural mesothelioma: a study based on The Cancer Genome Atlas database and experimental validation
Highlight box
Key findings
• We tentatively conclude that three prognostic marker genes [ubiquitin like with PHD and ring finger domain 1 (UHRF1), kinesin family member 4A (KIF4A), and never in mitosis gene A-related kinase 2 (NEK2)] as a gene cluster may serve as prognostic marker genes in pleural mesothelioma (PM).
What is known and what is new?
• Early detection and prognostic prediction are crucial in improving the survival of patients with PM.
• Our study uses molecular features as a breakthrough point to identify prognosis-related genes associated with PM.
What is the implication, and what should change now?
• Three prognostic marker genes (UHRF1, KIF4A, and NEK2) as a gene cluster may serve as prognostic marker genes in PM.
• More clinical samples were needed to validate the gene prognostic model that we have constructed.
Introduction
Pleural mesothelioma (PM) is an aggressive tumor on the surface of the pleura with a very poor prognosis after diagnosis and a median survival of less than 2 years (1). Over the past decade, its incidence and mortality rates have shown a yearly increasing trend globally. In many parts of Eastern Europe, Asia, South America, and Africa, where asbestos production and commercial usage are poorly regulated, the rates may continue to rise over the coming decades (1), indicating that even after reaching peak incidence, PM will remain a significant global health concern.
Moreover, the first-line treatment option for PM is pemetrexed combined with cisplatin or pemetrexed combined with cisplatin and bevacizumab (2). However, due to intra- and inter-tumor heterogeneity, as well as the presence of intrinsic and acquired drug resistance in tumor cells, the clinical benefits of chemotherapy or targeted therapy for PM patients remain very limited. Therefore, it is crucial to identify effective predictive biomarkers to enhance the efficacy of these treatment modalities, improve PM prognosis, and increase survival rates for PM patients.
Features used to predict the survival period of PM patients can generally be divided into clinical and molecular characteristics. Clinical features include clinical staging, pathological types, age at initial diagnosis, etc. (3); molecular features include gene mutations, gene expression characteristics, etc. (4). Research has found that clinical features have certain limitations in predicting survival in PM patients, indicating that current clinical prognostic factors are insufficient for accurately predicting individual clinical outcomes (5). This underscores the necessity of identifying highly sensitive, specific, and reliable prognostic biomarkers in PM. Despite recent progress in related research, the molecular basis of this disease largely remains unknown, which is the key limitation in developing effective targeted treatment strategies for PM (5).
With the advancement of bioinformatics and high-throughput sequencing technologies, gene expression features considering patients’ genetic and genomic differences have gradually become new tools for assessing cancer survival in humans (6). Since the molecular basis of PM is largely unknown and represents a key limitation for targeted treatment strategies, this study uses molecular features as a breakthrough point to identify prognosis-related genes associated with PM. The aim is to construct prognostic models to predict which patients might benefit the most from treatment.
Therefore, this study aims to use molecular features as a breakthrough for predicting survival in PM patients. By integrating data from multiple databases and employing a range of bioinformatics analysis methods, we identify and explore genes associated with PM prognosis as molecular features. Subsequently, we construct a 3-gene prognostic model as a prognostic biological marker that demonstrates strong predictive performance for PM. Additionally, we investigate the potential biological mechanisms of the prognostic model genes in the development and progression of PM. Ubiquitin like with PHD and ring finger domain 1 (UHRF1), kinesin family member 4A (KIF4A), and never in mitosis gene A-related kinase 2 (NEK2) in the prognostic model participate in multiple tumor-related signaling pathways and functions. Their expression is significantly correlated with various clinical characteristics of PM patients, suggesting their potential roles in the occurrence and development of PM. The findings of this research provide new insights for basic research on PM. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2531/rc).
Methods
Data acquisition and preprocessing
To establish a gene-related prognostic signature of PM, we downloaded RNA sequencing (RNA-seq) data and the corresponding clinicopathological information of 87 samples from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) (6). Due to a lack of information on normal lung tissues in the TCGA cohort, we also extracted data of normal lung samples from the Genotype-Tissue Expression (GTEx) database (https://www.gtexportal.org/home/). Gene expression levels were filtered with the selection standard of values greater than 1 in 50% of the samples and normalized. For clinical data information obtained from PM patients in the TCGA database, R programming language was employed to extract relevant information such as patient IDs, death time, follow-up status, etc., while removing any duplicate clinical information.
Integration of gene expression and survival data for PM patients and dataset partitioning
The clinical information and gene expression profiles were merged by Perl software (Version 0.9.6), samples missing survival time and survival status were deleted. The filter P value of gene survival analysis was set to 0.05, and the complete data set was randomly divided into training set and testing set by using the createDataPartition function in the caret package.
Construction of the prognostic risk model
Based on univariate Cox proportional hazards regression analysis, the relationship between gene expression and overall survival (OS) was analyzed using the “survival” R package in the training set, genes with P value <0.05 were considered statistically significant and were defined as target genes. To enhance the feasibility and reliability of clinically relevant prognostic genes, next, the genes that were highly relevant to prognosis were further selected from the target genes by performing robustness test using the Rbsurv package, and these genes were analyzed in a multivariate Cox regression analysis, finally their regression coefficients were weighted to establish a risk scoring formula.
Selection of the optimal threshold based on receiver operating characteristic (ROC) curve
ROC curve was obtained using the survival ROC package of R software, optimal cut-off point was selected with maximum sensitivity and specificity, the difference in survival between the low- and high-risk groups classified according to the optimal cut-off point was assessed by Kaplan-Meier curves, and compared using the Log-Rank test, and the risk scoring formula was further validated by fitting the testing set and the complete set.
Validation of the prognostic risk model
Based on the risk scoring formula, the risk score of each patient was calculated and the patients were divided into high-risk group and low-risk group, distribution of risk score, survival status, and microRNA (miRNA) expression were plotted to compare the difference in survival between high-risk group and low-risk group. Forest plots were drawn to evaluate the independent prediction ability of gene prognosis models. All of the above analyses were performed in R version 4.1.2 software, and P<0.05 was considered statistically significant.
Clinical sample collection
The clinical samples, including 41 PM tissues and their paired non-tumor tissues, were obtained from histopathologically diagnosed PM patients who underwent curative resection at The First Affiliated Hospital of Dali University. All PM patients provided written informed consent for their enrollment in this study, and the study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. This study has been approved by the Medical Ethics Committee of Dali University (No. 2020-YXLL22).
Cell culture
The human mesothelioma cell lines (NCI-H2452, NCI-H28, NCI-H2052, MTSO-211H) and mesothelial cell line (Met5A) used in this study were purchased from Pricella biotech Co., Ltd. (Wuhan, China) and Cellcook biotech Co., Ltd. (Guangzhou, China). All cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Grand Island, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, USA) at 37 ℃ in a humidified incubator containing 5% carbon dioxide. All cells were authenticated via short tandem repeat (STR) profiling (Pricella, Wuhan, China). Mycoplasma status was checked every month using the TransDetect Luciferase Mycoplasma Detection Kit (TransGen Biotech, Beijing, China). The medium was changed every 2–3 days, and 0.25% trypsin solution (Thermo Fisher HyClone, Utah, USA) was added for cell digestion and cell passage when the cells were 80% confluent.
The quantitative reverse transcription polymerase chain reaction (qRT-PCR) verification of genes in the prognostic model
According to manufacturer’s instructions, total RNA was isolated from the cell lines and tissues by using the Trelief RNAprep FastPure Tissue & Cell Kit (CAT No. TSP413, Tsingke, Beijing, China). Then, the RNA was transcribed into complementary DNA (cDNA) using HiScriptIIQ RT SuperMix for qPCR (+gDNA wiper) according to the manufacturer’s instructions (CAT No. R223-01, Vazyme, Nanjing, China), qRT-PCR was performed using 2×Universal Blue SYBR Green qPCR Master Mix (CAT No. G3326-1, Servicebio, Wuhan, China) on an ABI 7500 Real-Time polymerase chain reaction (PCR) system (Applied Biosystems, Waltham, MA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference, and the relative expression levels of the RNAs were calculated with the comparative threshold cycle (2−ΔΔCt) method. The experiment was conducted with four biological replicates and three technical replicates. The PCR primers used in this study are shown in Table 1.
Table 1
| Gene | Sequences (5'-3') |
|---|---|
| UHRF1 | F: CCACATCGTCCTCACAGC |
| R: GGTCCACATCATCCTCATAGC | |
| KIF4A | F: TCTGTTTCAGGCTGCTTTCA |
| R: GCCCTGAAATATTTGATTGGAG | |
| NEK2 | F: CGACGGTTAAACGGGGC |
| R: TACAGCAAGCAGCCCAATGA | |
| GAPDH | F: GAACGGGAAGCTCACTGG |
| R: CCTGCTTCACCACCTTCT |
F, forward primer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KIF4A, kinesin family member 4A; NEK2, never in mitosis gene A-related kinase 2; qRT-PCR, quantitative reverse transcription polymerase chain reaction; R, reverse primer; UHRF1, ubiquitin-like with PHD and ring finger domain 1.
Validation of gene expression in the prognostic model
The gene expression FPKM (fragments per kilo base of transcript per million mapped fragments) data and sample information file (GTEx phenotype) for normal lung tissue were downloaded from the GTEx database in the UCSC Xena database (http://xena.ucsc.edu/). Since the FPKM data in the GTEx database is transformed using log2(FPKM + 0.001), while the FPKM data in the TCGA database is transformed using log2(FPKM + 1), it is necessary to unify the two sets of FPKM data using a Perl script. Next, the expression matrix was subjected to ID conversion via Perl scripts to convert Ensembl IDs to corresponding gene names, and finally the GTEx and TCGA data were merged for further differential analysis for expression validation of prognostic model genes.
Analysis of gene expression patterns in PM within the prognostic model
The University of ALabama at birmingham CANcer data analysis portal (UALCAN) was a comprehensive and interactive web resource for analyzing cancer genomics data, providing easy access to publicly available cancer genomics data from TCGA as well as providing graphs depicting gene expression and patient survival information based on gene expression (7). After logging into the UALCAN database homepage (http://ualcan.path.uab.edu/), users selected TCGA for differential analysis of gene transcription levels, chose TCGA gene, entered the gene name in the option box under Enter gene symbol(s), selected the TCGA dataset for Mesothelioma, clicked Explore, selected Expression under Links for analysis, which output box plots of the expression patterns of genes in prognostic models in patients with PM, and analyzed the expression of genes in PM in these prognostic models.
Gene set enrichment analysis (GSEA) of genes in the prognostic model
GSEA is a bioinformatics method used to study whether specific gene sets (such as functional categories or pathways) are significantly enriched in gene expression data, helping to reveal potential mechanisms related to biological processes or diseases (8). PM samples from the TCGA database were divided into high- and low-expression groups based on the median expression of genes in the prognostic gene model. A difference analysis was performed on the two groups, and the genes were sorted from large to small according to the log fold change (FC) values of the treated genes to form a sorted gene list that represented the expression trend of the genes between the two groups. The sorted gene list was used as an input file for GSEA analysis, and the enrichment score (ES) of the gene set was calculated. The ES was finally defined as the largest peak. Next, a significance test was performed on the ES value of the gene set to estimate the significance level of the ES. Finally, multiple hypothesis testing was performed. The ES calculated for each gene subset was standardized according to the size of the gene set to obtain the normalized ES. The false positive rate was then calculated for the NES to identify the significantly enriched gene sets.
Statistical analysis
The GraphPad Prism software (version 8.0) and R software (version 4.0.2) were employed to process all statistical analyses. Quantitative data are expressed as the mean ± standard deviation (SD). Comparative analyses between two groups were performed utilizing either paired or unpaired Student’s t-test, contingent upon the normality of the data distribution and the equality of variances. P<0.05 was used as the threshold for significance in all analyses.
Results
Preprocessing of TCGA gene expression datasets
A total of 87 RNA-seq datasets of PM with corresponding clinical follow-up information are extracted from the public TCGA database. The PM patient expression profile containing 52,678 gene expression levels is obtained after filtering. The final expression level of a gene was defined as log2 (x + 1) of the original expression level. The R sofeware was used to perform normalization correction respectively and draw box plots of the gene expression distribution of each sample before and after the treatment (Figure 1). The gene expression levels of each sample after normalization tend to be consistent.
Using Cox univariate regression analysis and single-factor robust test to screen prognostic-related genes
According to the selected threshold, 30,433 genes expressed in samples from all patients were filtered out from a total of 52,678 genes. Further screening for genes with significant variation across different samples yielded a total of 24,350 genes. After removing samples with missing survival status and survival time, as well as duplicate samples, these 24,350 genes were further randomly divided into training and testing sets. The training set contained 43 samples, while the testing set contained 42 samples. Performing univariate Cox regression analysis on the training set, we evaluated the relationship between gene expression levels and patient OS, calculating each individual factor and screening out significant factors. This process identified 2,753 aberrantly expressed genes associated with OS (filtered using a threshold of P<0.05), which were further defined as target genes. To enhance the accuracy of the constructed model, single-factor robust test was conducted on target genes with significant expression identified in the univariate Cox regression analysis, resulting in the identification of 7 prognostic-related genes (Table 2).
Table 2
| Gene | nloglik | AIC |
|---|---|---|
| UHRF1 | 178.83 | 359.67* |
| CHAF1B | 176.62 | 357.24* |
| XRCC2 | 175.69 | 357.38* |
| CORO1C | 173.04 | 354.08* |
| KIF4A | 171.46 | 352.92* |
| NEK2 | 170.07 | 352.14* |
| LMNB2 | 168 | 349.99* |
| KPNA2 | 167.82 | 351.63 |
| MYBL2 | 167.73 | 353.46 |
| ECT2 | 167.67 | 355.34 |
| CDC25A | 167.37 | 356.75 |
| AURKA | 167.04 | 358.07 |
| CDC20 | 166.35 | 358.7 |
| TRIP13 | 166.33 | 360.65 |
| IQGAP3 | 166.25 | 362.5 |
| TOP2A | 166.07 | 364.15 |
| DEPDC1 | 165.92 | 365.83 |
| NCAPG | 162.94 | 361.88 |
| TROAP | 162.36 | 362.73 |
| CDCA3 | 161.63 | 363.26 |
| SGO1 | 160.29 | 362.58 |
| BIRC5 | 159.81 | 363.62 |
| GTSE1 | 159.81 | 365.61 |
| ANLN | 159.49 | 366.99 |
| JPT1 | 157 | 364.01 |
| CENPN | 156.67 | 365.33 |
| POLQ | 155.69 | 365.37 |
| KIF23 | 155.15 | 366.29 |
| CDCA2 | 152.76 | 363.53 |
*, P<0.05. AIC, Akaike information criterion.
Cox multivariable regression analysis for constructing PM prognostic model
Cox multivariable regression analysis was performed on these 7 prognostic genes, screen for significant factors (Table 3), and obtain the hazard ratio (HR) values and P values. Applying Cox multivariable regression analysis to reanalyze the results of these 7 genes, and finally select 3 significant genes along with their corresponding coefficients, and construct a multivariable model (Table 4). To comprehensively investigate the association between these 3 identified genes and PM prognosis, a risk score formula based on the Cox coefficients of these genes was established: risk score = UHRF1 expression level × 1.4525 − KIF4A expression level × 1.327 + NEK2 expression level × 1.4167.
Table 3
| Gene | coef | exp(coef) | se(coef) | z | Pr(>|z|) |
|---|---|---|---|---|---|
| UHRF1 | 0.9637 | 2.6213 | 0.4925 | 1.957 | 0.0504 |
| CHAF1B | 0.4398 | 1.5524 | 0.6425 | 0.685 | 0.4936 |
| XRCC2 | 0.9503 | 2.5866 | 0.6221 | 1.528 | 0.1266 |
| CORO1C | 0.7563 | 2.1303 | 0.3957 | 1.911 | 0.0560 |
| KIF4A | −2.0726 | 0.1259 | 0.8931 | −2.321 | 0.0203* |
| NEK2 | 1.4739 | 4.3662 | 0.7022 | 2.099 | 0.0358* |
| LMNB2 | 0.2657 | 1.3043 | 0.5344 | 0.497 | 0.6190 |
*, P<0.05. CHAF1B, chromatin assembly factor 1 subunit B; CORO1C, coronin 1C; KIF4A, kinesin family member 4A; LMNB2, lamin B2; NEK2, never in mitosis gene A-related kinase 2; UHRF1, ubiquitin-like with PHD and ring finger domain 1; XRCC2, X-ray repair cross complementing 2.
Table 4
| Gene | coef | exp(coef) | se(coef) | z | Pr(>|z|) |
|---|---|---|---|---|---|
| UHRF1 | 1.4525 | 4.2737 | 0.4255 | 3.414 | 0.000641*** |
| KIF4A | −1.327 | 0.2653 | 0.6802 | −1.951 | 0.051081 |
| NEK2 | 1.4167 | 4.1235 | 0.6558 | 2.16 | 0.030746* |
*, P<0.05; ***, P<0.001. KIF4A, kinesin family member 4A; NEK2, never in mitosis gene A-related kinase 2; UHRF1, ubiquitin-like with PHD and ring finger domain 1.
Selection of optimal threshold based on ROC curve
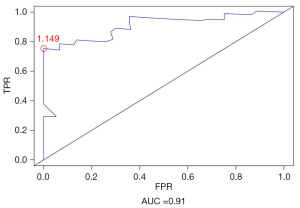
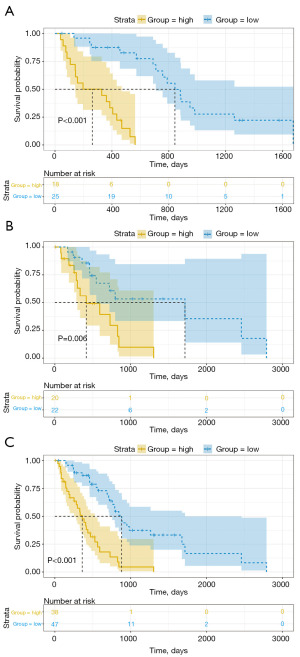
To compare the sensitivity and specificity of survival prediction, ROC analysis was performed for the 3-gene risk score model (Figure 2). Most cut-off points achieved good classification, with an area under the curve of 0.91. The optimal cutoff point was determined to be 1.149, which maximized sensitivity and specificity. At this optimal cutoff point, patients were further divided into high-risk (n=18) and low-risk (n=25) groups. Kaplan-Meier curves and log-rank tests further indicated a significant difference in survival time between the high- and low-risk groups (P<0.001; Figure 3A). To assess the accuracy of the risk model in prognosis prediction, the risk model was applied to the testing set and the complete dataset to evaluate the accuracy of the model in PM prognosis prediction. Using the selected optimal cutoff point, patients in testing set were classified into high- (n=20) and low-risk (n=22) groups, consistent with the results of the training set. The Kaplan-Meier curves of the two groups further showed that the survival time of low-risk patients was significantly prolonged, while the survival time of high-risk patients was significantly shortened (P=0.006) (Figure 3B). Patients in the complete dataset were classified into high- (n=38) and low-risk (n=47) groups based on the selected optimal cutoff point. Although there was an imbalance in the number of samples between the groups, similar results were obtained from survival analysis, indicating significantly prolonged survival time for low-risk patients compared to high-risk patients (P<0.001; Figure 3C).
Validation of the prognostic gene model for PM
The cumulative risk plot reflects the cumulative death risk magnitude of the study subjects at a certain time point, with the horizontal axis representing survival time and the vertical axis representing the cumulative risk (HR) of events occurring over time (Figure 4). In the training set, test set, and entire dataset, at the same time point, high-risk patients have a greater cumulative death risk compared to low-risk patients, indicating a lower probability of surviving beyond that time point, which also demonstrates the significant discriminatory effect of the constructed predictive model.

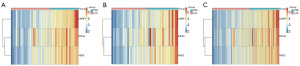
Based on the correlation coefficient in the risk regression model and corresponding gene expression levels, the risk score for each patient is calculated. Scatter plots of patient risk score distribution, survival status, and high and low-risk heatmaps of the expression levels of the three prognostic genes are plotted for the training set, testing set, and complete set to visualize the analysis results (Figures 5-7). The results indicate that with an increase in risk score, patients exhibit a trend of shortened survival time, and the proportion of deaths is higher in the high-risk group compared to the low-risk group, with the number of deceased patients gradually increasing. Therefore, patients in the high-risk group have shorter survival times and higher mortality rates. The heatmaps show that UHRF1, KIF4A, and NEK2 exhibit a gradually increasing expression trend from left to right. From the scatter plots of patient risk score distribution as well as the high and low-risk heatmaps, we observed that UHRF1, KIF4A, and NEK2 exhibit a positive correlation with risk score.


The PM gene prognostic risk model is visualized using a forest plot (Figure 8). This forest plot divides the genes associated with the PM prognostic model into high- and low-risk groups based on X=1, where the right side represents high-risk and the left side represents low-risk. In this model, a total of 2 genes are located to the right of this dashed line, and these 2 genes (UHRF1, NEK2) are statistically significant (P<0.05), indicating their role as independent predictors of high risk for PM prognosis. Additionally, one gene is located to the left of the dashed line, which is not statistically significant (P>0.05), indicating its role as a low-risk predictor for PM prognosis. The overall Log-Rank P<0.001 for the entire model, indicating a good predictive ability.
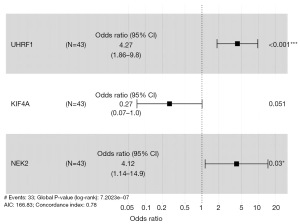
Schoenfeld residuals test was conducted to assess the independence of residuals with respect to time, thereby testing the proportional hazards assumption in the Cox model. The results are shown in the figure. The horizontal axis represents time, the residuals are evenly distributed, and residual and time are independent of each other. The P values are all greater than 0.1 (PUHRF1=0.32, PKIF4A=0.11, PNEK2=0.57), indicating that the respective variables in the entire model meet the prerequisite of proportional hazards (Figure 9).

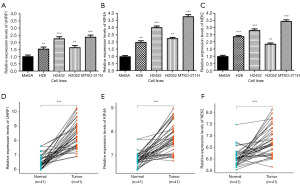
Detection of UHRF1, KIF4A, and NEK2 expression in cell lines and clinical samples
UHRF1, KIF4A, and NEK2 expression levels were measured in human cell lines and clinical samples using qRT-PCR. In the human mesothelioma cell lines (NCI-H2452, NCI-H28, NCI-H2052, MTSO-211H), the expression levels of UHRF1, KIF4A, and NEK2 were significantly increased, comparing with mesothelial cell line (Met5A) (Figure 10A-10C). Furthermore, UHRF1, KIF4A, and NEK2 levels were consistent with the results obtained in cell lines, the three genes expression were still significantly upregulated in PM tissues, comparing with non-tumor tissues, indicating that UHRF1, KIF4A, and NEK2 promotes tumorigenesis (Figure 10D-10F). Thus, owing to its consistently high expressions, we speculate that UHRF1, KIF4A, and NEK2 may promotes the development and progression of PM.
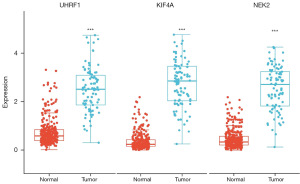
Validation of differential gene expression in the prognostic model
Further exploration of gene expression in the prognostic model was conducted. Firstly, we validated whether the expression level of the three genes, UHRF1, KIF4A, and NEK2, in the prognostic model exhibited differential expression in a total of 374 samples from the TCGA and GTEx databases. Box plots were utilized for visualization. The results indicated significant differential expression of these three genes in PM tumor tissues compared to normal lung tissues (P<0.001), with all three genes showing higher expressions in tumor tissues (Figure 11).
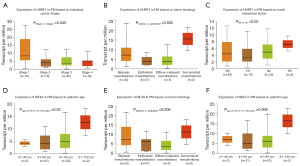
Expression patterns of genes in PM in the prognostic model
Next, to explore the expression patterns of prognostic model genes in PM patients, we utilized the UALCAN database to generate box plots illustrating the expression patterns of three prognostic model genes, UHRF1, KIF4A, and NEK2, across tumor stage, gender, age, tumor subtype, and lymph node metastasis status in PM patients. The expression of these genes in PM was analyzed, and the results were visualized with statistical significance (Figure 12). The results show that UHRF1 exhibits significant expression differences across different PM stages (P=0.045), which can distinguish between Stage 1 and Stage 2 of PM. UHRF1 also shows significant expression differences across different PM tumor subtypes (P=0.003), which can distinguish between biphasic and epithelioid subtypes. UHRF1 expression also significantly differs across different lymph node metastasis statuses (P=0.02), which can distinguish between N0 and N1 statuses. KIF4A shows significant expression differences across different age groups in PM patients (P=0.02), with significant differences between the age groups of 41–60 and 81–100 years. KIF4A also exhibits significant expression differences across different tumor subtypes in PM patients, which can distinguish between biphasic and epithelioid subtypes (P=0.006). NEK2 demonstrates significant expression differences across different age groups in PM patients, with significant differences between the age groups of 41–60 and 81–100 years (P=0.008). Therefore, the roles of these genes in the progression of PM are worth further investigation. In addition, correlation analysis between UHRF1, KIF4A, and NEK2 showed significant positive correlations between the three genes (Figure S1).
GSEA of genes in the prognostic model
Subsequently, to explore the potential biological functions of genes in the prognostic model, we employed GSEA to investigate enrichment pathways associated with gene expression in the prognostic model. Samples were stratified into high and low expression groups based on the median expression values of UHRF1, KIF4A, and NEK2 genes in PM patients from the TCGA dataset. Enrichment analysis was conducted using R software. The results revealed that the high-expression group of UHRF1 was significantly enriched in pathways related to DNA replication, proteasomes, and homologous recombination. The high-expression group of KIF4A showed significant enrichment in pathways related to DNA replication and proteasomes, while the high-expression group of NEK2 was significantly enriched in pathways related to DNA replication and nucleocytoplasmic transport. These findings suggest potential biological functions of these genes in PM (Figure 13). All three high-expression groups of these genes were significantly enriched in pathways related to DNA replication, Fanconi anemia pathway, messenger RNA (mRNA) surveillance pathway, nucleocytoplasmic transport, ribosome biogenesis in eukaryotes, and spliceosome pathway.

Discussion
PM is a rare malignant tumor originating from the mesothelial cells of the pleura. Due to its long latency period and nonspecific symptoms, patients have very poor prognosis, with a 5-year OS rate of 5% (9). Asbestos exposure is a major risk factor for PM (10). However, a small proportion of PM cases appear unrelated to asbestos exposure (11). Recent studies on PM tumorigenesis have highlighted the role of certain tumor-associated genes, which can influence the development and prognosis of PM (11,12). At the moment, while there is considerable research on the diagnosis and treatment of PM, there remains a lack of methods for evaluating prognosis and effectively reducing recurrence and mortality rates. Therefore, there is an urgent need for research on new methods and highly sensitive and specific biomarkers to assess the prognosis of PM. In recent years, the combination of surgery and chemotherapy, as well as optimization of chemotherapy, has improved the survival and quality of life of PM patients (12). However, long-term survival in PM patients is rare, making it difficult to determine which biological factors can clearly distinguish between patients with poor progression-free survival (PFS) and OS and those with improved outcomes. Therefore, tumor biomarkers are receiving increasing attention due to their minimal invasiveness. Exploring biomarkers for PM can aid in screening and early diagnosis, thereby improving prognosis.
In this study, we downloaded gene expression profiles and corresponding PM clinical information from the TCGA database. After integrating all gene expression information with clinical survival data, we constructed a prognostic model consisting of three genes, UHRF1, KIF4A, and NEK2, using single-factor Cox regression, single-factor robust test screening analysis, and multi-factor Cox regression methods. The risk scores calculated by this model not only predict patient prognosis well in the training set but also show significant differences between high- and low-risk groups in the testing set and the complete dataset. Additionally, the risk score of this model can serve as an independent factor influencing prognosis in the TCGA dataset, indicating that this model can serve as a prognostic biological marker for PM patients.
Further exploration of gene expression in the prognostic model was conducted. Validation of UHRF1, KIF4A, and NEK2 was performed using the TCGA and GTEx databases, revealing significant differences in expression between PM tumor tissues and normal lung tissues, with all three genes showing high expression in tumor tissues. Additionally, upon deeper investigation into the expression patterns of genes in the prognostic model, UHRF1, KIF4A, and NEK2 were found to exhibit significant differences in expression across different stages, tumor subtypes, ages, and metastatic states of PM, suggesting their potential roles in PM progression. GSEA enrichment analysis of these three genes revealed significant enrichment in pathways related to DNA replication, Fanconi anemia pathway, mRNA surveillance pathway, nuclear transport, ribosome biogenesis in eukaryotes, and spliceosome pathway, all of which are associated with mechanisms underlying tumor development. Thus, GSEA enrichment analysis elucidated the potential biological functions of high expression of these three genes in PM occurrence and progression.
Furthermore, increased expression of the three genes was observed in vitro in cell lines and paired clinical samples. We further investigated the research status of these genes by searching relevant literature. UHRF1 is a regulator of DNA methylation (13-15) and a driver of epigenetic inheritance in various human cancers (14,16). Recent reports have indicated its overexpression in mesothelioma (17). Immunohistochemical staining of UHRF1 can distinguish benign reactive spindle cell mesothelial proliferation from sarcomatoid mesothelioma (18). In small cell lung cancer, UHRF1 is highly expressed in tumor tissue (19). Elevated levels of UHRF1 indicate a poorer prognosis in small cell lung cancer. UHRF1 binds to YAP1 and inhibits YAP1 ubiquitination degradation, thereby stabilizing YAP1 protein in small cell lung cancer cells (19). Downregulation of UHRF1 can enhance the sensitivity of small cell lung cancer cells to cisplatin and is associated with favorable prognosis in small cell lung cancer patients treated with platinum-based chemotherapy (19). Therefore, UHRF1 can serve as a biomarker for predicting the prognosis of small cell lung cancer patients and as a potential therapeutic target for small cell lung cancer patients. Luo and colleagues’ current research has identified UHRF1 as a significant regulatory factor in breast cancer growth (20). Clinical data analysis reveals that compared to normal breast tissue, UHRF1 is significantly elevated in breast cancer, and its expression is associated with poor survival rates in breast cancer patients (20). UHRF1 is located in the nucleus and interacts with estrogen receptor alpha (ERα) through its SET and RING associated (SRA) domain, thereby inhibiting the K48 ubiquitination modification of ERα and enhancing ERα stability (20). Therefore, UHRF1 is highly expressed in cancers such as lung cancer and breast cancer and is associated with poor prognosis. It is likely a potential predictive biomarker and therapeutic target for PM, and further mechanistic research in PM is warranted.
KIF4A is a member of the kinesin superfamily of motor proteins, primarily located in the cell nucleus, and its function within the nucleus vary with the cell cycle stages. Human KIF4A is a novel component of the chromosome condensation and segregation machinery functioning in multiple steps of mitotic division (21,22). Some studies suggest that KIF4A is involved in DNA damage and repair processes, and abnormal expression of KIF4A may affect the expression of the homologous recombination enzyme RadS1 and its regulator BRCA2, leading to failure in repairing damaged DNA (23,24). DNA damage may result in abnormal cell proliferation and differentiation, ultimately promoting tumor formation (25). Therefore, KIF4A is closely associated with the occurrence and prognosis of various human tumors (26). Research suggests that KIF4A is significantly correlated with OS in patients with cholangiocarcinoma, with higher expression of KIF4A being associated with lower survival rates. There is a significant correlation between KIF4A and infiltration of activated memory T cells and activated mast cells in the tumor microenvironment. Increased expression of KIF4A affects the degree of immune cell infiltration, which may be involved in the regulation of immune tolerance by cholangiocarcinoma cells (27). KIF4A can also regulate the biological functions of esophageal squamous cell carcinoma cells through the Hippo signaling pathway, promoting the proliferation and migration of esophageal squamous cell carcinoma cells. High expression of KIF4A is associated with poor OS in esophageal squamous cell carcinoma (28). As a result, KIF4A is closely associated with the occurrence and prognosis of various human tumors such as cholangiocarcinoma and esophageal squamous cell carcinoma. As an oncogene in multiple cancers, based on this study and previous research, KIF4A is likely an effective predictive biomarker for PM, and further exploration in PM is needed.
NEK2 is one of the members of the never in mitosis gene A (NIMA)-related kinase family. It is a multifunctional protein involved in cell cycle regulation, such as centrosome replication and separation, microtubule stabilization, centromere attachment, and spindle assembly checkpoint (29). Aberrant expression of NEK2 and dysregulated downstream protein phosphorylation mediated by NEK2 are frequently observed to be associated with the occurrence, progression, and metastasis of tumors, leading to poorer prognosis in various cancers (29). The research by Zhang et al. demonstrates that NEK2 phosphorylates programmed cell death ligand 1 (PD-L1) to maintain its stability, resulting in poor efficacy of PD-L1-targeted immunotherapy in pancreatic cancer. They identified NEK2 as an immune-related prognostic factor for pancreatic cancer, which participates in the development of pancreatic tumors in an immune-dependent manner. Lack of NEK2 leads to suppression of PD-L1 expression and enhanced lymphocyte infiltration (29). Wan et al. investigated the relationship between NEK2 and gastric cancer, and found that the progression of gastric cancer is associated with overexpression of NEK2, especially in patients with larger tumor volumes and lymph node metastasis. Their research indicates that overexpression of NEK2 binds to and inhibits protein phosphatase 1 (PP1), subsequently activating protein kinase B (AKT) and downstream oncogenic pathways. Through the AKT/hypoxia-inducible factor-1 alpha (HIF1α) axis, glucose metabolism is reprogrammed into aerobic glycolysis, providing rapid energy for the growth of gastric cancer cells. Furthermore, inhibition of autophagy activity through the AKT/mammalian target of rapamycin (mTOR) axis results in reduced efficacy of cancer treatment, promoting tumor cell survival. Conversely, silencing NEK2 to deactivate AKT will decrease aerobic glycolysis, promote autophagic cell death, and ultimately inhibit the growth of gastric cancer cells (30). Consequently, NEK2 has an oncogenic role in various cancers such as pancreatic cancer and gastric cancer, influencing multiple tumor-related pathways. NEK2 is likely an effective therapeutic target and predictive biomarker for PM, and its specific mechanistic role in PM warrants further investigation.
Early detection of cancers by genes relies primarily on the detection of tumor-associated gene mutations, abnormal expression, or epigenetic changes. Liquid biopsy enables non-invasive early tumor screening by analyzing genetic information in circulating tumor DNA (ctDNA), circulating tumor cells (CTCs) or exosomes in the blood. ctDNA testing detects genetic mutations, such as point mutations, insertions/deletions, copy number variations, etc., by performing high-throughput sequencing of DNA fragments released into the blood by tumor cells. The CTC assay analyzes CTCs in the blood for gene expression or mutations. Exosome assays can be used to analyze RNA or proteins in exosomes secreted by tumor cells to identify tumor markers. The three genes in this model have been confirmed to be highly expressed in cells and tissues through experimental and bioinformatics analysis, and whether there are abnormalities in the expression levels in body fluids and their regulatory mechanisms still need to be further explored.
Conclusions
In conclusion, these three genes play significant roles in tumor formation and development. However, their specific molecular mechanisms in PM are less reported, suggesting potential directions for future research. The prognostic model we constructed can calculate the mortality risk of individual patients through the expression of genes, and help doctors judge the prognosis of patients, so as to formulate personalized treatment and interventions. There are limitations in this study that need to be addressed, due to the significant benefit that early-stage PM patients can derive from clinical intervention, this model fails to effectively distinguish high-risk patients among early-stage PM cases. Therefore, the model cannot provide guidance for the development of more effective clinical intervention measures, such as implementing more thorough treatment protocols or shortening review intervals. This aspect requires further improvement in the future, and we need more clinical samples to validate the gene prognostic model that we have constructed.
Acknowledgments
We greatly appreciate the hard work of our editors and reviewers.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2531/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2531/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2531/prf
Funding: This research received financial support from
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2531/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. This study has been approved by the Medical Ethics Committee of The First Affiliated Hospital of Dali University (No. 2020-YXLL22). All PM patients provided written informed consent for their enrollment in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sinn K, Mosleh B, Hoda MA. Malignant pleural mesothelioma: recent developments. Curr Opin Oncol 2021;33:80-6. [Crossref] [PubMed]
- Yang H, Xu D, Schmid RA, et al. Biomarker-guided targeted and immunotherapies in malignant pleural mesothelioma. Ther Adv Med Oncol 2020;12:1758835920971421. [Crossref] [PubMed]
- Carbone M, Adusumilli PS, Alexander HR Jr, et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J Clin 2019;69:402-29. [Crossref] [PubMed]
- Dubois F, Bazille C, Levallet J, et al. Molecular Alterations in Malignant Pleural Mesothelioma: A Hope for Effective Treatment by Targeting YAP. Target Oncol 2022;17:407-31. [Crossref] [PubMed]
- Yap TA, Aerts JG, Popat S, et al. Novel insights into mesothelioma biology and implications for therapy. Nat Rev Cancer 2017;17:475-88. [Crossref] [PubMed]
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Shavelle R, Vavra-Musser K, Lee J, et al. Life Expectancy in Pleural and Peritoneal Mesothelioma. Lung Cancer Int 2017;2017:2782590. [Crossref] [PubMed]
- Cinausero M, Rihawi K, Cortiula F, et al. Emerging therapies in malignant pleural mesothelioma. Crit Rev Oncol Hematol 2019;144:102815. [Crossref] [PubMed]
- Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 2011;43:1022-5. [Crossref] [PubMed]
- Campbell NP, Kindler HL. Update on malignant pleural mesothelioma. Semin Respir Crit Care Med 2011;32:102-10. [Crossref] [PubMed]
- Bronner C, Alhosin M, Hamiche A, et al. Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns. Genes (Basel) 2019;10:65. [Crossref] [PubMed]
- Sidhu H, Capalash N. UHRF1: The key regulator of epigenetics and molecular target for cancer therapeutics. Tumour Biol 2017;39:1010428317692205. [Crossref] [PubMed]
- Zhang H, Liu H, Chen Y, et al. A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat Commun 2016;7:10201. [Crossref] [PubMed]
- Ashraf W, Ibrahim A, Alhosin M, et al. The epigenetic integrator UHRF1: on the road to become a universal biomarker for cancer. Oncotarget 2017;8:51946-62. [Crossref] [PubMed]
- Reardon ES, Shukla V, Xi S, et al. UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma. J Thorac Oncol 2021;16:89-103. [Crossref] [PubMed]
- Yang H, Cheung S, Churg A. UHRF1 Immunohistochemical Staining Separates Benign Reactive Spindle Cell Mesothelial Proliferations From Sarcomatoid Mesotheliomas. Am J Surg Pathol 2022;46:840-5. [Crossref] [PubMed]
- Hou J, Li W, Zhang S, et al. UHRF1 plays an oncogenic role in small cell lung cancer. Mol Carcinog 2023;62:385-97. [Crossref] [PubMed]
- Luo G, Li Q, Yu M, et al. UHRF1 modulates breast cancer cell growth via estrogen signaling. Med Oncol 2022;39:111. [Crossref] [PubMed]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, et al. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J 2004;23:3237-48. [Crossref] [PubMed]
- Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol 2004;166:613-20. [Crossref] [PubMed]
- Lee YM, Kim W. Association of human kinesin superfamily protein member 4 with BRCA2-associated factor 35. Biochem J 2003;374:497-503. [Crossref] [PubMed]
- Wu G, Zhou L, Khidr L, et al. A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle 2008;7:2013-20. [Crossref] [PubMed]
- Taniwaki M, Takano A, Ishikawa N, et al. Activation of KIF4A as a prognostic biomarker and therapeutic target for lung cancer. Clin Cancer Res 2007;13:6624-31. [Crossref] [PubMed]
- Narayan G, Bourdon V, Chaganti S, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer 2007;46:373-84. [Crossref] [PubMed]
- Zhang DY, Ma SS, Sun WL, et al. KIF4A as a novel prognostic biomarker in cholangiocarcinoma. Medicine (Baltimore) 2021;100:e26130. [Crossref] [PubMed]
- Sun X, Chen P, Chen X, et al. KIF4A enhanced cell proliferation and migration via Hippo signaling and predicted a poor prognosis in esophageal squamous cell carcinoma. Thorac Cancer 2021;12:512-24. [Crossref] [PubMed]
- Zhang X, Huang X, Xu J, et al. NEK2 inhibition triggers anti-pancreatic cancer immunity by targeting PD-L1. Nat Commun 2021;12:4536. [Crossref] [PubMed]
- Wan H, Xu L, Zhang H, et al. High expression of NEK2 promotes gastric cancer progression via activating AKT signaling. J Physiol Biochem 2021;77:25-34. [Crossref] [PubMed]


