
Prognostic significance of IL11RA and its methylation in lung adenocarcinoma
Introduction
According to the data of 36 cancers from 185 countries in the world, lung cancer is still the leading cause of cancer morbidity and mortality. There are about 2.48 million new cases and 1.81 million deaths every year (1). Over 40% of all malignant tumors in the lungs are lung adenocarcinoma (LUAD), which is the most frequent pathological type (2). For patients with early-stage LUAD, surgery is an effective treatment that ensures a good prognosis and a high quality of life (3). For those with locally advanced or advanced LUAD who are not eligible for surgery, treatment options include radiotherapy, chemotherapy, immunotherapy, and targeted therapy (4). Even with these therapeutic advancements, there has been little improvement in the 5-year survival rate of LUAD (5). Hence, it is of profound significance to identify new biomarkers for forecasting LUAD prognosis and underlying therapeutic targets.
Interleukin-11 receptor alpha (IL11RA), a specific receptor for interleukin-11 (IL-11), is differentially expressed in various malignancies, like stomach cancer, prostate cancer, breast cancer, endometrial carcinoma, osteosarcoma, melanoma, and colon cancer (6-9). Currently, IL11RA is identified as being related to tumor progression, cell proliferation, and differentiation, implying its potential involvement in the process of carcinogenesis (10,11). Interestingly, IL11RA is markedly decreased in LUAD (12). But it is unclear whether low expression of IL11RA acts as a tumor suppressor or pro-oncogenic effect in LUAD. One recent study demonstrated that IL11/IL11RA signaling pathways may contribute to the immunomodulatory properties (13). Higher levels of IL11RA expression enhanced immune cell infiltration and suppressed tumor growth (14). Nevertheless, the link between IL11RA and immune microenvironment in LUAD is not well comprehended. Hence, this research aimed to elucidate the differential expression, methylation, prognosis value and immune infiltration of IL11RA in LUAD. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2362/rc).
Methods
Acquisition of patient data
According to our previous research (15), transcriptomic data from The Cancer Genome Atlas (TCGA), including 59 normal and 526 LUAD tissue samples, were sourced from the University of California Santa Cruz (UCSC) Xena website (http://xena.ucsc.edu/). The transcriptomic data comprising 59 normal and 513 LUAD subjects were included in subsequent analysis after eliminating duplicate samples. Additionally, the UCSC Xena website also provided DNA methylation data for 32 normal and 471 LUAD subjects. For the meta-analysis, expression and survival data were sourced from TCGA and Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) databases (16). The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
The IL11RA expression analysis in LUAD
According to our previous research (17), we examined IL11RA expression across various cancer types using the University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN) (http://ualcan.path.uab.edu/) (18), Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) (19) and ONCOMINE (https://www.oncomine.org/) (20) databases. as well as the TCGA and GEO databases. Paired t-test and unpaired t-test was employed to assess the differences in IL11RA expression between the groups. Moreover, IL11RA protein expression levels were evaluated through immunohistochemistry in the Human Protein Atlas (HPA) database (http://www.proteinatlas.org) (21). To explore the connection between IL11RA expression levels and clinicopathological characteristics, we excluded some missing data and performed chi-square tests. We also estimated the expression of IL11RA under diverse clinical pathological parameters by Student’s t-test or Kruskal-Wallis test (22).
The IL11RA methylation analysis in LUAD
The methylation level of IL11RA in LUAD was evaluated by UALCAN database. After excluding patients who survive <30 days, survival differences among groups with distinct methylation levels were evaluated using the Kaplan-Meier (KM) approach (23). Likewise, to explore the connection between the distinct methylation levels of IL11RA and clinicopathological characteristics, we excluded some missing data and performed chi-square tests.
The prognostic analysis of IL11RA in LUAD
The data retrieval entry of meta-analysis was as follows in GEO database: (lung OR pulmonary OR respiratory OR bronchi OR bronchioles OR alveoli OR pneumocytes OR (air way)) AND (cancer OR tumor OR carcinoma OR neoplasm OR malignant OR adenocarcinoma) AND (survival) (24). Two thousand nine hundred and sixty-four LUAD patients from TCGA and 22 GEO data set were included for meta-analysis (25-45). According to our previous research (46), the “survminer” and “survival” packages in R software were utilized to individually calculate the hazard ratio (HR) and 95% confidence intervals (CIs) for 23 datasets. Then, a meta-analysis was performed through “meta” packages in R software. Random effect model was selected based on I2>50% and P<0.05 (47). Additionally, we also created a prognostic nomogram model using “regplot” R packages in R software for LUAD patients based on IL11RA gene expression and clinicopathologic characteristics, like age, gender, stage, T stage, N stage and M stage.
Gene set enrichment analysis (GSEA)
To clarify the possible mechanism of IL11RA in LUAD, GSEA was performed in LUAD patients of TCGA datasets through “clusterProfiler” packages in R software according to our previous research (48). All samples were divided into high expression group and low expression group according to the median value of IL11RA expression.
The correlation analysis between IL11RA expression and immune infiltration in LUAD
To evaluate the relevance between IL11RA expression and immune infiltration in LUAD, according to our previous research (49,50), we firstly used the “Estimate” R package to calculate immune, stromal, and ESTIMATE scores. Afterward, the “Gene Set Variation Analysis (GSVA)” R package was used to execute single sample gene set enrichment analysis (ssGSEA) to better illustrate the connection between IL11RA expression and 28 types of tumor-infiltrating immune cells (51). Finally, Tumor Immune Estimation Resource (TIMER; https://cistrome.shinyapps.io/timer/) was utilized to assess the association of IL11RA expression with B cells in LUAD and the survival difference between the high and low B cell. Besides, TIMER was used to analyze the association between IL11RA expression and immune cell markers.
Statistical analysis
R software facilitated all the statistical computations. The link between IL11RA expression and its DNA methylation levels was examined using the Spearman method. P<0.05 was regarded to be statistically different. Gene sets were deemed statistically distinct if their adjust P value <0.05.
Results
The expression of IL11RA is diminished in LUAD
To appraise the IL11RA expression in diverse malignancies, we performed the expression analysis of IL11RA in pan-cancer through UALCAN database (Figure 1A). The results we obtained indicated that compared with that in the normal tissues, the IL11RA messenger RNA (mRNA) expression were considerably increased in only liver hepatocellular carcinoma, while it was markedly decreased in 15 other cancers compared to normal tissue, including LUAD. The pan-cancer analysis from ONCOMINE database also demonstrated that IL11RA expression is down-regulated in the majority of tumors (Figure 1B). There is an indication that IL11RA could function as a tumor suppressor gene. In order to further validate the distinct expression of IL11RA in normal and LUAD samples, we performed the expression analysis based on multiple databases. The findings revealed that the mRNA expression of IL11RA in LUAD tissues from GEPIA (Figure 1C), TCGA (Figure 1D), GSE75037 (Figure 1E) and GSE31210 (Figure 1F) databases were significantly reduced compared with normal tissue. Furthermore, representative images of immunohistochemistry results from HPA database showed that the staining intensity of IL11RA was moderate positivity in LUAD tissues, whereas strong positivity in normal samples (Figure 1G). The findings indicate that IL11RA may exert significant roles in the tumorigenesis of LUAD.

Analysis of IL11RA methylation profile in LUAD patients
It is well established that hypermethylation results in gene silencing. Consequently, we speculate whether DNA hypermethylation leads to down-regulation of IL11RA expression in LUAD. According to the UALCAN database, DNA methylation levels of IL11RA was considerably elevated in LUAD samples relative to normal samples (Figure 2A). IL11RA mRNA expression levels were found to be negatively correlated with DNA methylation levels in the TCGA databases (Figure 2B). The hypermethylation of IL11RA implied inferior prognosis in LUAD (Figure 2C). Above results indicate that hypermethylation negatively regulates the IL11RA gene, leading to its reduced expression in LUAD. Furthermore, we also explored the link between IL11RA expression and 5 methylation sites in the promoter region of IL11RA, Figure 2D clearly showed the distribution of 5 methylation sites. The results of correlation analysis illustrated that IL11RA expression is negatively linked with 4 methylation sites (Figure 2E). It is especially intriguing that methylation sites cg21504624 (Figure 2F) and cg14609668 (Figure 2G) was inversely related to the expression of IL11RA in LUAD, and their hypermethylation was closely linked to unfavorable overall survival (Figure 2H,2I). The findings imply that the hypermethylation of IL11RA may lead to its downregulation in LUAD, contributing to a worse prognosis for subjects.
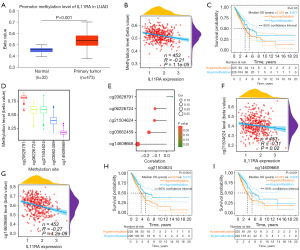
The relevance between IL11RA expression or its methylation and clinical pathological parameters of LUAD
We next explored the relevance between IL11RA expression or its methylation and clinical pathological parameters from the TCGA-LUAD project using the chi-square test. Our findings suggested that the expression of IL11RA was tightly linked with gender (P=0.009), stage (P=4.38e−04), T stage (P=3.77e−05), N stage (P=0.003), cancer status (P=5.76e−04) and IL11RA methylation (P=9.64e−05), while the methylation level of IL11RA was tightly linked with stage (P=0.03), T stage (P=0.02), N stage (P=0.03), primary therapy outcome (P=0.007) and IL11RA mRNA expression (P=4.36e−05) (Table 1). Interestingly, further examination indicated a reduction in IL11RA expression as the stage (Figure 3A, P<0.001), T stage (Figure 3B, P<0.001), and N stage (Figure 3C, P<0.001) advances, whereas no noteworthy link between IL11RA expression and other clinicopathologic characteristics, like M stage (Figure 3D, P=0.68), age (Figure 3E, P=0.72) and gender (Figure 3F, P=0.17). Besides, those with tumors (a patient with LUAD still has a tumor in his body after treatment) (Figure 3G, P=0.003) or new tumor event (new tumor grows in a lung cancer patient after treatment) (Figure 3H, P=0.003) exhibited much lower levels of IL11RA expression.
Table 1
| Clinical features | IL11RA expression | IL11RA methylation | |||||
|---|---|---|---|---|---|---|---|
| High, n (%) | Low, n (%) | P value | High (%) | Low (%) | P value | ||
| Age (years) | 0.47 | >0.99 | |||||
| ≤65 | 115 (46.37) | 123 (50.00) | 114 (50.22) | 114 (50.67) | |||
| >65 | 133 (53.63) | 123 (50.00) | 113 (49.78) | 111 (49.33) | |||
| Gender | 0.009 | 0.75 | |||||
| Female | 153 (59.77) | 123 (47.86) | 123 (52.34) | 128 (54.24) | |||
| Male | 103 (40.23) | 134 (52.14) | 112 (47.66) | 108 (45.76) | |||
| Race | 0.22 | 0.42 | |||||
| White/American | 192 (85.33) | 207 (85.89) | 179 (84.04) | 182 (87.50) | |||
| Black/Africa American | 31 (13.78) | 27 (11.20) | 29 (13.62) | 24 (11.54) | |||
| Asian | 2 (0.89) | 7 (2.90) | 5 (2.35) | 2 (0.96) | |||
| Stage | 4.38e−04 | 0.03 | |||||
| I | 157 (62.06) | 117 (46.43) | 117 (50.21) | 142 (60.94) | |||
| II | 59 (23.32) | 62 (24.60) | 65 (27.90) | 49 (21.03) | |||
| III | 27 (10.67) | 57 (22.62) | 44 (18.88) | 29 (12.45) | |||
| IV | 10 (3.95) | 16 (6.35) | 7 (3.00) | 13 (5.58) | |||
| T stage | 3.77e−05 | 0.02 | |||||
| T1 | 108 (42.52) | 60 (23.44) | 69 (29.36) | 90 (38.63) | |||
| T2 | 123 (48.43) | 153 (59.77) | 142 (60.43) | 109 (46.78) | |||
| T3 | 17 (6.69) | 30 (11.72) | 20 (8.51) | 22 (9.44) | |||
| T4 | 6 (2.36) | 13 (5.08) | 4 (1.70) | 12 (5.15) | |||
| N stage | 0.003 | 0.03 | |||||
| N0 | 181 (72.69) | 149 (59.13) | 143 (61.11) | 166 (73.78) | |||
| N1 | 44 (17.67) | 51 (20.24) | 50 (21.37) | 34 (15.11) | |||
| N2 | 23 (9.24) | 51 (20.24) | 40 (17.09) | 25 (11.11) | |||
| N3 | 1 (0.40) | 1 (0.40) | 1 (0.43) | 0 (0.00) | |||
| M stage | 0.40 | 0.30 | |||||
| M0 | 161 (94.71) | 183 (91.96) | 158 (95.76) | 147 (92.45) | |||
| M1 | 9 (5.29) | 16 (8.04) | 7 (4.24) | 12 (7.55) | |||
| Cancer status | 5.76e−04 | 0.23 | |||||
| Tumor-free | 135 (72.58) | 108 (55.10) | 111 (60.33) | 120 (66.67) | |||
| With tumor | 51 (27.42) | 88 (44.90) | 73 (39.67) | 60 (33.33) | |||
| Kras mutation | 0.64 | 0.47 | |||||
| Yes | 10 (32.26) | 12 (41.38) | 10 (34.48) | 12 (48.00) | |||
| No | 21 (67.74) | 17 (58.62) | 19 (65.52) | 13 (52.00) | |||
| Radiation | 0.26 | >0.99 | |||||
| Yes | 31 (14.69) | 24 (10.67) | 23 (11.68) | 23 (11.56) | |||
| No | 180 (85.31) | 201 (89.33) | 174 (88.32) | 176 (88.44) | |||
| Primary therapy outcome | 0.76 | 0.007 | |||||
| CR | 143 (71.50) | 143 (74.48) | 139 (79.43) | 135 (71.81) | |||
| PR | 3 (1.50) | 4 (2.08) | 6 (3.43) | 0 (0.00) | |||
| SD | 15 (7.50) | 15 (7.81) | 11 (6.29) | 18 (9.57) | |||
| PD | 39 (19.50) | 30 (15.62) | 19 (10.86) | 35 (18.62) | |||
| KPS | 0.87 | 0.92 | |||||
| ≤80 | 14 (42.42) | 17 (47.22) | 18 (47.37) | 14 (51.85) | |||
| >80 | 19 (57.58) | 19 (52.78) | 20 (52.63) | 13 (48.15) | |||
| IL11RA messenger RNA expression | – | 4.36e−05 | |||||
| High | – | – | 92 (40.17) | 134 (59.82) | |||
| Low | – | – | 137 (59.83) | 90 (40.18) | |||
| IL11RA methylation | 9.64e−05 | – | |||||
| High | 93 (40.61) | 133 (59.38) | – | – | |||
| Low | 136 (59.39) | 91 (40.62) | – | – | |||
CR, complete response; IL11RA, interleukin-11 receptor alpha; KPS, Karnofsky performance scale; LUAD, lung adenocarcinoma; PD, progressive disease; PR, partial response; SD, stable disease; TCGA, The Cancer Genome Atlas.

Reduced IL11RA expression correlates with a negative prognosis in LUAD
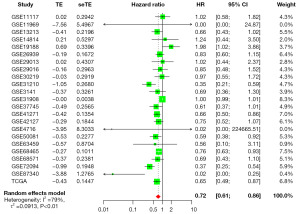
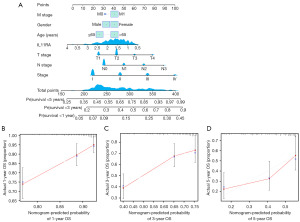
To elucidate the relevance between IL11RA expression and prognosis in LUAD, a meta-analysis was performed based on TCGA and GEO databases. Two thousand nine hundred and sixty-four patients from 23 studies were incorporated into this meta-analysis (Table 2). Of these 23 studies, 18 had HR less than 1 and 5 had HR equal or greater than 1. The pooled hazard ratios for the correlation between differentially expressed IL11RA and overall survival was 0.72 (95% CI: 0.61–0.86) (Figure 4), which indicates that low IL11RA expression is linked with deteriorated overall survival in LUAD. Furthermore, a nomogram for prognosis was designed to enhance the accuracy of survival predictions for LUAD patients based on IL11RA gene expression and clinicopathologic characteristics, like age, gender, stage, T stage, N stage and M stage. The results indicated that IL11RA gene expression and stage had the largest impact on prognosis prediction (Figure 5A). Further calibration curves confirmed that the nomogram’s predictions aligned well with the actual survival probabilities observed at 1-, 3-, and 5-year (Figure 5B-5D).
Table 2
| ID | Contributor [year] | Experimental platform | Number of LUAD |
|---|---|---|---|
| GSE11117 | Baty F [2010] (25) | GPL6650 | 13 |
| GSE11969 | Takeuchi T [2006] (26) | GPL7015 | 90 |
| GSE13213 | Tomida S [2009] (27) | GPL6480 | 117 |
| GSE14814 | Zhu CQ [2010] (28) | GPL96 | 71 |
| GSE19188 | Hou J [2010] (29) | GPL570 | 40 |
| GSE26939 | Wilkerson MD [2012] (30) | GPL9053 | 115 |
| GSE29013 | Xie Y [2011] (31) | GPL570 | 30 |
| GSE29016 | Staaf J [2012] (32) | GPL6947 | 38 |
| GSE30219 | Rousseaux S [2013] (33) | GPL570 | 85 |
| GSE31210 | Okayama H [2012] (34) | GPL570 | 226 |
| GSE3141 | Bild AH [2006] (35) | GPL570 | 58 |
| GSE31908 | Girard L [2011] | GPL570 | 20 |
| GSE37745 | Botling J [2013] (36) | GPL570 | 106 |
| GSE41271 | Sato M [2013] (37) | GPL6884 | 182 |
| GSE42127 | Tang H [2013] (38) | GPL6884 | 133 |
| GSE4716 | Tomida S [2004] (39) | GPL3694 | 30 |
| GSE50081 | Der SD [2014] (40) | GPL570 | 127 |
| GSE63459 | Robles AI [2015] (41) | GPL6883 | 33 |
| GSE68465 | Shedden K [2008] (42) | GPL96 | 442 |
| GSE68571 | Beer DG [2002] (43) | GPL80 | 83 |
| GSE72094 | Schabath MB [2016] (44) | GPL15048 | 398 |
| GSE87340 | Sun Z [2014] (45) | GPL11154 | 27 |
| TCGA | – | – | 500 |
| Total | – | – | 2,964 |
LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
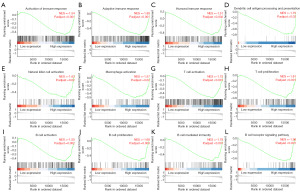
IL11RA is involved in immune infiltration in LUAD
To shed light on the underlying biological roles of IL11RA in LUAD, we carried out GSEA analysis. Interestingly, the results demonstrated that multiple significant pathways related to immune response were enriched in the IL11RA high expression group (Figure 6A-6L), such as activation of immune response (P.adjust<0.001), natural killer cell activation (P.adjust=0.02), macrophage activation (P.adjust=0.02), T cell activation (P.adjust<0.001), B cell activation (P.adjust=0.002) and so on, which suggests that IL11RA might be implicated in tumor immune infiltration.
Next, we measured the immune, stromal, and ESTIMATE scores to evaluate the immune status in the high versus low IL11RA expression groups. The analysis revealed that the group with reduced IL11RA expression had lower scores for immune (P<0.001), stromal (P=0.03), and ESTIMATE (Figure 7A, P<0.001). Above results reveal that IL11RA mRNA expression might be positively correlated with tumor immune infiltration. Moreover, we also carried out ssGSEA analysis to better clarity and concise present the link between IL11RA expression and tumor-infiltrating immune cells. As shown in Figure 7B, 17 of 28 immune cells were notably under-expressed in low IL11RA expression groups, whereas 4 immune cells exhibited elevated expression in low IL11RA expression groups. Findings from the correlation analysis demonstrated that IL11RA mRNA expression were significantly positively related to 17 of 28 immune cells, negatively linked with 4 immune cells, and had no statistically significant correlation with 10 immune cells (Figure 7C). What is intriguing is that both our analysis and the TIMER database results demonstrated a positive relationship between IL11RA expression and B cell infiltration (Figure 7D-7F), and the survival analysis results demonstrated that patients with LUAD who showed higher B cell expression experienced longer survival according to the TIMER database (Figure 7G). Above results suggests that IL11RA low expression may exert cancer-promoting effect by influencing the immune cells infiltration in LUAD, especially B cell.
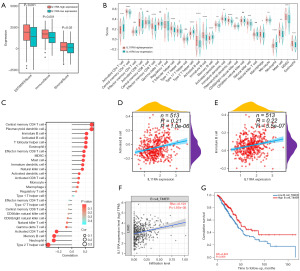
Ultimately, we utilized the TIMER database to analyze the link between IL11RA expression and markers of immune cells. The study discovered that 44 out of 52 immune cell markers, or 84.62%, showed a notable connection with IL11RA expression. Of the 52 correlations, 76.92% were positive, totaling 40, and 7.69% were negative, totaling 4 (Table 3). Above findings reveal a significant relationship between IL11RA expression and most immune cell markers in LUAD.
Table 3
| Description | Gene markers | None adjusted | Tumor purity adjusted | |||
|---|---|---|---|---|---|---|
| Cor | P value | Cor | P value | |||
| Activated B cell | CD19 | 0.21 | 1.52e−06 | 0.226 | 4.11e−07 | |
| CD79A | 0.105 | 0.02 | 0.101 | 0.03 | ||
| GNG7 | 0.456 | 8.78e−28 | 0.456 | 1.02e−26 | ||
| BLK | 0.301 | 3.09e−12 | 0.323 | 1.88e−13 | ||
| CLEC9A | 0.242 | 2.73e−08 | 0.232 | 1.96e−07 | ||
| Immature B cell | CD22 | 0.388 | 5.62e−20 | 0.425 | 4.57e−23 | |
| FCRL1 | 0.329 | 1.9e−14 | 0.349 | 1.52e−15 | ||
| FAM129C | 0.345 | 7.45e−16 | 0.374 | 9.05e−18 | ||
| TXNIP | 0.284 | 5.19e−11 | 0.281 | 2.01e−10 | ||
| STAP1 | 0.276 | 1.98e−10 | 0.277 | 4.02e−10 | ||
| Memory B cell | FCER1A | 0.309 | 6.95e−13 | 0.305 | 4.39e−12 | |
| SOX5 | 0.228 | 1.67e−07 | 0.232 | 1.99e−07 | ||
| TLR9 | 0.137 | 0.002 | 0.134 | 0.003 | ||
| CCNA2 | −0.408 | 4.95e−22 | −0.42 | 1.53e−22 | ||
| CDKN3 | −0.389 | 4.39e−20 | −0.407 | 4.03e−21 | ||
| Th1 | T-bet (TBX21) | 0.199 | 5.07e−06 | 0.19 | 2.15e−05 | |
| STAT4 | 0.231 | 1.19e−07 | 0.221 | 6.95e−07 | ||
| TNF-a (TNF) | 0.17 | 1.01e−04 | 0.148 | 9.77e−04 | ||
| STAT1 | −0.082 | 0.06 | −0.125 | 0.005 | ||
| Th2 | STAT6 | 0.257 | 3.15e−09 | 0.269 | 1.22e−09 | |
| STAT5A | 0.278 | 1.3e−10 | 0.27 | 1.11e−09 | ||
| IL13 | 0.165 | 1.67e−04 | 0.144 | 0.001 | ||
| GATA3 | 0.104 | 0.02 | 0.072 | 0.11 | ||
| Tfh | BCL6 | 0.236 | 6.11e−08 | 0.232 | 1.89e−07 | |
| IL21 | −0.042 | 0.34 | −0.069 | 0.13 | ||
| Th17 | STAT3 | 0.129 | 0.003 | 0.126 | 0.005 | |
| IL17A | −0.063 | 0.15 | −0.07 | 0.12 | ||
| Treg | STAT5B | 0.329 | 1.89e−14 | 0.333 | 3.26e−14 | |
| FOXP3 | 0.135 | 0.002 | 0.12 | 0.008 | ||
| CCR8 | 0.084 | 0.057 | 0.063 | 0.16 | ||
| T cell exhaustion | CTLA4 | 0.147 | 7.97e−04 | 0.12 | 0.008 | |
| TIM-3 (HAVCR2) | 0.12 | 0.007 | 0.082 | 0.07 | ||
| GZMB | −0.186 | 2.25e−05 | −0.243 | 4.76e−08 | ||
| PD-1 (PDCD1) | 0.073 | 0.10 | 0.045 | 0.32 | ||
| CD8+ T cell | CD8A | 0.043 | 0.34 | 0.013 | 0.77 | |
| CD8B | 0.055 | 0.22 | 0.031 | 0.49 | ||
| M1 macrophage | IRF5 | 0.213 | 1.02e−06 | 0.195 | 1.27e−05 | |
| INOS (NOS2) | 0.058 | 0.19 | 0.044 | 0.33 | ||
| COX2 (PTGS2) | −0.094 | 0.03 | −0.115 | 0.01 | ||
| M2 macrophage | CD163 | 0.103 | 0.02 | 0.063 | 0.16 | |
| VSIG4 | 0.143 | 0.001 | 0.112 | 0.01 | ||
| MS4A4A | 0.156 | 3.72e−04 | 0.123 | 0.006 | ||
| Neutrophil | CD66b (CEACAM8) | 0.278 | 1.33e−10 | 0.279 | 2.7e−10 | |
| CD11b (ITGAM) | 0.256 | 3.89e−09 | 0.234 | 1.54e−07 | ||
| CCR7 | 0.271 | 3.9e−10 | 0.289 | 6.16e−11 | ||
| Dendritic cell | HLA-DPB1 | 0.382 | 2.36e−19 | 0.39 | 2.35e−19 | |
| HLA-DPA1 | 0.326 | 3.13e−14 | 0.326 | 1.15e−13 | ||
| BDCA-1 (CD1C) | 0.329 | 1.95e−14 | 0.323 | 2.07e−13 | ||
| CD11C (ITGAX) | 0.315 | 2.45e−13 | 0.308 | 2.84e−12 | ||
| TAM | IL10 | 0.17 | 1.04e−04 | 0.144 | 0.001 | |
| CCL2 | 0.075 | 0.09 | 0.045 | 0.32 | ||
| CD68 | 0.098 | 0.03 | 0.071 | 0.11 | ||
IL11RA, interleukin-11 receptor alpha; TAM, tumor-associated macrophages; Tfh, T follicular helper cells; Th1, T helper cell 1; Th2, T helper cell 2; Th17, T helper cell 17; TIMER, Tumor Immune Estimation Resource; Treg, T regulatory cells.
Discussion
The occurrence and progression of malignant tumors are caused by the interaction and influence of numerous factors and genes (52,53). Research indicates a strong link between lung cancer progression and irregular gene expression (54). Hence, it is of great significance to explore the pathogenesis and prognosis of lung cancer at the gene level, and to find new molecular mechanisms and effective therapeutic targets for LUAD. In this research, we demonstrated that IL11RA is lowly expressed and negatively correlated with its methylation level in LUAD. The prognosis of patients with low expression of IL11RA is poor, and the mechanism may be related to the reduction of immune cell infiltration.
In cancer, IL11RA, as a cell surface receptor, has been discovered to act as a protein that is both secretory and bound to membranes (55). Research has previously indicated that IL11RA is strongly linked to the development of tumors (56). In many types of neoplasm tissues, the IL11RA gene showed aberrant expression. In this research, we clarified that IL11RA mRNA expression was markedly diminished in the majority of tumors, including LUAD. The result implies that IL11RA may act as a potential tumor suppressor gene. In addition, the significant reduction in mRNA and protein expression of IL11RA in LUAD was confirmed by several databases. This tendency of low expression of IL11RA in LUAD coincides with previous research (12), but some tumors, like those in the stomach and breast, show inconsistency (9,57). This discrepancy may be associated with small sample sizes of normal tissues or incongruent detection assays. Interestingly, wo also observed a reduction in IL11RA expression as the stage, T stage, and N stage advances, which indicates that IL11RA may play a significant role in the cancer development and progression of LUAD. Our result suggests that IL11RA could be considered a promising molecular target for LUAD therapy.
Then, we carried out a prognostic meta-analysis of 2,964 LUAD patients from 23 studies based on TCGA and GEO databases. Of these 23 studies, 18 had HR less than 1 and 5 had HR equal or greater than 1. However, the pooled hazard ratios for the correlation between differentially expressed IL11RA and overall survival was less than 1. The findings indicated that lower IL11RA expression correlated with poorer outcomes in LUAD. These findings effectively highlighted that IL11RA might be a viable prognostic biomarker for LUAD.
Abnormal DNA methylation plays a role in the development and advancement of LUAD (58). By modulating the expression of oncogenes and tumor suppressor genes, DNA methylation contributes to the onset and growth of tumors (59,60). Thus, we speculate whether the low expression of IL11RA in LUAD is associated with DNA methylation? In this research, there was an inverse relationship between IL11RA mRNA expression and its methylation in LUAD. Furthermore, the hypermethylation of IL11RA implied dismal overall survival in LUAD. This result indicates that IL11RA gene is inversely regulated by hypermethylation, resulting in its low expression in LUAD.
Another significant finding of the research was that IL11RA expression was linked with immune infiltration levels in LUAD. In lung cancer, the tumor microenvironment is mainly populated by T cells, with B cells, macrophages, dendritic cells, and natural killer cells also being significant (61). By facilitating the movement of immune cells near malignant tumors, immune activating factors contribute to anti-tumor activity (62). Research has demonstrated that IL11RA and its related signaling pathways can significantly alter the tumor microenvironment and immune responses (14). Our findings suggested that multiple significant pathways related to immune response were enriched in the IL11RA high expression group, and the group with reduced IL11RA expression had lower scores for immune, stromal, and ESTIMATE, and IL11RA expression was closely linked to diverse immune cells, including activated CD4+ T cells, activated B cells, activated dendritic cells, natural killer cells, activated CD8+ T cells and so on. Moreover, IL11RA expression is also significantly linked with majority of immune cell markers in LUAD. Studies predominantly indicate that dendritic cells aid in the activation of antitumor T lymphocytes (63,64), and cancer patients with elevated T cell and B cell levels had improved survival outcomes (65-67). Further analysis illustrated that IL11RA expression was positively associated with B cell infiltration and LUAD patients with higher expression of B cells presented a longer survival time. Taken together, these findings imply that IL11RA could aid in the recruitment of immune infiltrating cells and may be involved in modulating the immune response in the development of LUAD, which leads to a less favorable survival outcome for LUAD patients.
Conclusions
In conclusion, our study suggests that IL11RA gene is inversely regulated by hypermethylation, resulting in its low expression in LUAD and decreased IL11RA expression is positively related to dismal survival outcome. Our work highlights that IL11RA may be a viable prognostic biomarker and an innovative molecular therapeutic target for individuals with LUAD. Nevertheless, we initially investigated the biological function of IL11RA in LUAD by bioinformatics analysis. Additional biomedical experiments are required to clarify the concrete molecular mechanism that links low IL11RA expression to the progression of LUAD.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2362/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2362/prf
Funding: This investigation was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2362/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Li H, Sha X, Wang W, et al. Identification of lysosomal genes associated with prognosis in lung adenocarcinoma. Transl Lung Cancer Res 2023;12:1477-95. [Crossref] [PubMed]
- Xing P, Wang S, Cao Y, et al. Treatment strategies and drug resistance mechanisms in adenocarcinoma of different organs. Drug Resist Updat 2023;71:101002. [Crossref] [PubMed]
- Chen J, Shao J, Zhang X, et al. Monitoring early dynamic changes of plasma cell-free DNA and pretreatment pre-albumin to predict chemotherapy effectiveness and survival outcomes in advanced non-small cell lung cancer. Ann Transl Med 2022;10:253. [Crossref] [PubMed]
- Du Z, Zhang F, Liu L, et al. LncRNA ANRIL promotes HR repair through regulating PARP1 expression by sponging miR-7-5p in lung cancer. BMC Cancer 2023;23:130. [Crossref] [PubMed]
- Zhang ZG, Zhu HM, Hua HK. Inhibiting IL11RA to mitigate hepatic metastasis in skin cutaneous melanoma: Comprehensive insights from in vitro and in vivo investigations. Skin Res Technol 2024;30:e13618. [Crossref] [PubMed]
- Yap J, Salamonsen LA, Jobling T, et al. Interleukin 11 is upregulated in uterine lavage and endometrial cancer cells in women with endometrial carcinoma. Reprod Biol Endocrinol 2010;8:63. [Crossref] [PubMed]
- Onnis B, Fer N, Rapisarda A, et al. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J Clin Invest 2013;123:1615-29. [Crossref] [PubMed]
- Calcagno DQ, Takeno SS, Gigek CO, et al. Identification of IL11RA and MELK amplification in gastric cancer by comprehensive genomic profiling of gastric cancer cell lines. World J Gastroenterol 2016;22:9506-14. [Crossref] [PubMed]
- Nakayama T, Yoshizaki A, Izumida S, et al. Expression of interleukin-11 (IL-11) and IL-11 receptor alpha in human gastric carcinoma and IL-11 upregulates the invasive activity of human gastric carcinoma cells. Int J Oncol 2007;30:825-33.
- Yoshizaki A, Nakayama T, Yamazumi K, et al. Expression of interleukin (IL)-11 and IL-11 receptor in human colorectal adenocarcinoma: IL-11 up-regulation of the invasive and proliferative activity of human colorectal carcinoma cells. Int J Oncol 2006;29:869-76.
- Gao L, Zhang L. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for lung adenocarcinoma. BMC Cancer 2021;21:849. [Crossref] [PubMed]
- Huynh J, Baloyan D, Chisanga D, et al. Host IL11 Signaling Suppresses CD4(+) T cell-Mediated Antitumor Responses to Colon Cancer in Mice. Cancer Immunol Res 2021;9:735-47. [Crossref] [PubMed]
- Ur Rehman A, Wang Z, Qin Q, et al. Enhancing antitumor immunity and achieving tumor eradication with IL11RA mRNA immunotherapy. Int Immunopharmacol 2024;134:112205. [Crossref] [PubMed]
- Wu YJ, Nai AT, He GC, et al. DPYSL2 as potential diagnostic and prognostic biomarker linked to immune infiltration in lung adenocarcinoma. World J Surg Oncol 2021;19:274. [Crossref] [PubMed]
- Wang Y, Liang S, Hong Q, et al. Construction of a neutrophil extracellular trap formation-related gene model for predicting the survival of lung adenocarcinoma patients and their response to immunotherapy. Transl Lung Cancer Res 2024;13:3407-25. [Crossref] [PubMed]
- Nai A, Zeng H, Wu Q, et al. lncRNA/miR-29c-Mediated High Expression of LOX Can Influence the Immune Status and Chemosensitivity and Can Forecast the Poor Prognosis of Gastric Cancer. Front Cell Dev Biol 2021;9:760470. [Crossref] [PubMed]
- Wan M, Meng H, Li H. Potential role of TWIST1 and its methylation in bladder urothelial carcinoma. Transl Cancer Res 2024;13:6070-86. [Crossref] [PubMed]
- Tang C, Li L, Zhu C, et al. GPR137-RAB8A activation promotes ovarian cancer development via the Hedgehog pathway. J Exp Clin Cancer Res 2025;44:22. [Crossref] [PubMed]
- Zheng Q, Xie Y, Xu L, et al. LDHA as a predictive biomarker and its association with the infiltration of immune cells in pancreatic adenocarcinoma. J Gastrointest Oncol 2024;15:1746-59. [Crossref] [PubMed]
- Wang H, Wang Y, Tan P, et al. Prognostic value and anti-tumor immunity role of TMED9 in pan-cancer: a bioinformatics study. Transl Cancer Res 2024;13:5429-45. [Crossref] [PubMed]
- Zhu X, Chen D, Sun Y, et al. LncRNA WEE2-AS1 is a diagnostic biomarker that predicts poor prognoses in patients with glioma. BMC Cancer 2023;23:120. [Crossref] [PubMed]
- Tan Y, Zhang S, Xiao Q, et al. Prognostic significance of ARL9 and its methylation in low-grade glioma. Genomics 2020;112:4808-16. [Crossref] [PubMed]
- Liang CY, Li ZY, Gan TQ, et al. Downregulation of hsa-microRNA-204-5p and identification of its potential regulatory network in non-small cell lung cancer: RT-qPCR, bioinformatic- and meta-analyses. Respir Res 2020;21:60. [Crossref] [PubMed]
- Baty F, Facompré M, Kaiser S, et al. Gene profiling of clinical routine biopsies and prediction of survival in non-small cell lung cancer. Am J Respir Crit Care Med 2010;181:181-8. [Crossref] [PubMed]
- Takeuchi T, Tomida S, Yatabe Y, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 2006;24:1679-88. [Crossref] [PubMed]
- Tomida S, Takeuchi T, Shimada Y, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol 2009;27:2793-9. [Crossref] [PubMed]
- Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol 2010;28:4417-24. [Crossref] [PubMed]
- Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 2010;5:e10312. [Crossref] [PubMed]
- Wilkerson MD, Yin X, Walter V, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One 2012;7:e36530. [Crossref] [PubMed]
- Xie Y, Xiao G, Coombes KR, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res 2011;17:5705-14. [Crossref] [PubMed]
- Staaf J, Jönsson G, Jönsson M, et al. Relation between smoking history and gene expression profiles in lung adenocarcinomas. BMC Med Genomics 2012;5:22. [Crossref] [PubMed]
- Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med 2013;5:186ra66. [Crossref] [PubMed]
- Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 2012;72:100-11. [Crossref] [PubMed]
- Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439:353-7. [Crossref] [PubMed]
- Botling J, Edlund K, Lohr M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res 2013;19:194-204. [Crossref] [PubMed]
- Sato M, Larsen JE, Lee W, et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res 2013;11:638-50. [Crossref] [PubMed]
- Tang H, Xiao G, Behrens C, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013;19:1577-86. [Crossref] [PubMed]
- Tomida S, Koshikawa K, Yatabe Y, et al. Gene expression-based, individualized outcome prediction for surgically treated lung cancer patients. Oncogene 2004;23:5360-70. [Crossref] [PubMed]
- Der SD, Sykes J, Pintilie M, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol 2014;9:59-64. [Crossref] [PubMed]
- Robles AI, Arai E, Mathé EA, et al. An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma Based on mRNA, microRNA, and DNA Methylation Biomarkers. J Thorac Oncol 2015;10:1037-48. [Crossref] [PubMed]
- Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7. [Crossref] [PubMed]
- Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816-24. [Crossref] [PubMed]
- Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209-16. [Crossref] [PubMed]
- Sun Z, Wang L, Eckloff BW, et al. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics 2014;7:32. [Crossref] [PubMed]
- Nai A, Ma F, He Z, et al. Development and Validation of a 7-Gene Inflammatory Signature Forecasts Prognosis and Diverse Immune Landscape in Lung Adenocarcinoma. Front Mol Biosci 2022;9:822739. [Crossref] [PubMed]
- He S, Shi J, Mao J, et al. The expression of miR-375 in prostate cancer: A study based on GEO, TCGA data and bioinformatics analysis. Pathol Res Pract 2019;215:152375. [Crossref] [PubMed]
- He JY, Li ZM, Chen YT, et al. Development and validation of a prognostic prediction model for cervical cancer patients treated with radical radiotherapy: a study based on TCGA database. Transl Cancer Res 2024;13:1721-36. [Crossref] [PubMed]
- Mo X, Hu D, Yang P, et al. A novel cuproptosis-related prognostic lncRNA signature and lncRNA MIR31HG/miR-193a-3p/TNFRSF21 regulatory axis in lung adenocarcinoma. Front Oncol 2022;12:927706. [Crossref] [PubMed]
- Mo X, Hu D, Li Y, et al. A novel pyroptosis-related prognostic lncRNAs signature, tumor immune microenvironment and the associated regulation axes in bladder cancer. Front Genet 2022;13:936305. [Crossref] [PubMed]
- Lee J, Yoon JH, Lee E, et al. Immune response and mesenchymal transition of papillary thyroid carcinoma reflected in ultrasonography features assessed by radiologists and deep learning. J Adv Res 2024;62:219-28. [Crossref] [PubMed]
- Jagga B, Edwards M, Pagin M, et al. Structural basis for nuclear import selectivity of pioneer transcription factor SOX2. Nat Commun 2021;12:28. [Crossref] [PubMed]
- Liu X, Yang S, Wang L, et al. Hierarchically tumor-activated nanoCRISPR-Cas13a facilitates efficient microRNA disruption for multi-pathway-mediated tumor suppression. Theranostics 2023;13:2774-86. [Crossref] [PubMed]
- Kong K, Hu S, Yue J, et al. Integrative genomic profiling reveals characteristics of lymph node metastasis in small cell lung cancer. Transl Lung Cancer Res 2023;12:295-311. [Crossref] [PubMed]
- Rehman AU, Olof Olsson P, Khan N, et al. Identification of Human Secretome and Membrane Proteome-Based Cancer Biomarkers Utilizing Bioinformatics. J Membr Biol 2020;253:257-70. [Crossref] [PubMed]
- Thanindratarn P, Dean DC, Nelson SD, et al. Chimeric antigen receptor T (CAR-T) cell immunotherapy for sarcomas: From mechanisms to potential clinical applications. Cancer Treat Rev 2020;82:101934. [Crossref] [PubMed]
- Irawan C, Atmakusumah D, Siregar NC, et al. Expression of Biomarkers CXCR4, IL11-RA, TFF1, MLF1P in Advanced Breast Cancer Patients with Bone Metastatic: a Diagnostic Study. Acta Med Indones 2016;48:261-8.
- Li R, Yang YE, Yin YH, et al. Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers. J Transl Med 2019;17:324. [Crossref] [PubMed]
- Niu W, Yang Y, Teng Y, et al. Pan-Cancer Analysis of PGAM1 and Its Experimental Validation in Uveal Melanoma Progression. J Cancer 2024;15:2074-94. [Crossref] [PubMed]
- Wang Y, Huo L, Yang C, et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and gastric cancer susceptibility: an updated meta-analysis. Biosci Rep 2023;43:BSR20222553. [Crossref] [PubMed]
- Pan M, Wang Y, Wang Z, et al. The mitosis-related gene OIP5 is a potential biomarker in pan-cancer. Ann Transl Med 2023;11:117. [Crossref] [PubMed]
- Li Y, Qi J, Yang J. RTP4 is a novel prognosis-related hub gene in cutaneous melanoma. Hereditas 2021;158:22. [Crossref] [PubMed]
- Chen F, Tang C, Yang F, et al. HSP90 inhibition suppresses tumor glycolytic flux to potentiate the therapeutic efficacy of radiotherapy for head and neck cancer. Sci Adv 2024;10:eadk3663. [Crossref] [PubMed]
- Le Naour J, Kroemer G. Trial watch: Toll-like receptor ligands in cancer therapy. Oncoimmunology 2023;12:2180237. [Crossref] [PubMed]
- Edlund K, Madjar K, Mattsson JSM, et al. Prognostic Impact of Tumor Cell Programmed Death Ligand 1 Expression and Immune Cell Infiltration in NSCLC. J Thorac Oncol 2019;14:628-40. [Crossref] [PubMed]
- Wang SS, Liu W, Ly D, et al. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol 2019;16:6-18. [Crossref] [PubMed]
- Zhang J, Endres S, Kobold S. Enhancing tumor T cell infiltration to enable cancer immunotherapy. Immunotherapy 2019;11:201-13. [Crossref] [PubMed]


