
The high expression of Long noncoding RNA TEX41 promotes the proliferation, migration, and invasion of hepatocellular carcinoma
Highlight box
Key findings
• Long noncoding RNA (lncRNA) TEX41 may promote hepatocellular carcinoma (HCC) progression by competitively binding to miR-200a-3p, leading to elevated BIRC5 expression, cell proliferation, migration, and invasion in HCC cells.
What is known and what is new?
• LncRNA TEX41 was overexpressed in HCC.
• LncRNA TEX41 promoted HCC progression, which may be achieved by targeting miR-200a-3p to regulate the expression of BIRC5.
What is the implication, and what should change now?
• In this study, we found that TEX41 may regulate HCC progression through the miR-200a-3p-BIRC5 axis. Further investigation of this regulatory mechanism may provide us with assistance in finding new liver cancer treatment methods.
Introduction
After lung cancer and colorectal cancer, liver cancer is the third leading cause of cancer death worldwide, with over 750,000 liver cancer-related deaths occurring in 2022 (1). Hepatocellular carcinoma (HCC) is the primary type of malignant liver disease. The 5-year survival rate for patients with primary localized HCC is 32.6%, while that for patients with regional or metastatic HCC is only 10.8% and 2.4%, respectively (2). Therefore, thoroughly examining the pathogenesis of HCC and finding new biomarkers have important guiding significance for determining treatment plans and improving prognosis. In recent studies, long noncoding RNA (lncRNA) has played an important role in the development of many cancers (3,4). Although lncRNA and microRNA (miRNA) cannot encode proteins, they also play an important role in regulating gene expression and protein function as noncoding transcripts (5). The mechanism of action of lncRNA includes gene imprinting, chromatin remodeling, cell cycle regulation, splicing regulation, messenger RNA degradation and translation regulation, and protein binding. Among them, lncRNA can adsorb miRNA through complementary base sequences, thereby acting as a competing endogenous RNA (ceRNA) to interact with miRNA and affect target gene expression. LncRNA TEX41 is a newly discovered lncRNA, and studies have confirmed its high expression and molecular sponge effect in various malignancies including cervical cancer, acute lymphoblastic leukemia, melanoma, lung adenocarcinoma, and colorectal cancer (6-8). Studies have shown that miR-200a-3p plays an important role in the emergence and development of HCC (9,10). BIRC5 is an antiapoptotic protein that participates in various physiological and pathological processes. Studies on this subject have shown that the high expression of BIRC5 increases the mortality rate of patients with HCC (11), and it may thus be valuable for diagnosing and treating this disease (12-14). We speculated that lncRNA TEX41 may target the miR-200a-3p-BIRC5 molecular axis and affect the development of HCC. In this study, we identified the binding sites between lncRNA TEX41 and miR-200a-3p, miR-200a-3p, and BIRC5 through the starBase database (https://rnasysu.com/encori/index.php). Subsequently, we sought to clarify the effects of lncRNA TEX41 on the biological behavior of HCC cells and the miR-200a-3p-BIRC5 molecular axis, providing a basis for the targeted therapy of HCC. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-812/rc).
Methods
Patients and samples
We retrospectively collected cancer tissues and adjacent tissues from patients with HCC who underwent surgery at The Second Affiliated Hospital of Nantong University from September 2022 to December 2024. The inclusion criteria were as follows: (I) meeting the diagnostic criteria for HCC and (II) successful acquisition and intact preservation of HCC and adjacent tissue specimens during operation. Meanwhile, the exclusion criteria were as follows: (I) a history of liver surgery; (II) malignant tumors in other parts of the body; (III) incomplete case data; and (IV) presence of autoimmune diseases. The basic clinical data of all patients with HCC, such as age, gender, tumor diameter, and tumor-node-metastasis (TNM) staging, were collected. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. This study was approved by the Medical Ethics Committee of The Second Affiliated Hospital of Nantong University (No. 2024KT186). Informed consent was taken from all individual participants.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA was separated with TRIzol reagent (15596026; Thermo Fisher Scientific, Waltham, MA, USA), and cDNA was prepared using the Takara Primer RT kit (RR036A; Takara Bio, Kusatsu, Japan), and quantitative polymerase chain reaction (qPCR) was conducted using BeyoFastTM SYBR Green qPCR Mix (D7262; Beyotime, Nantong, China) and a Bio-Rad CFX96 real-time fluorescence qPCR instrument (Bio-Rad Laboratories, California, USA). The qPCR conditions were as follows: 95 ℃ for 15 minutes, followed by 40 cycles, with each cycle consisting of 95 ℃ for 10 seconds and 66 ℃ for 32 seconds. Table 1 summarizes the primer sequences (Sangon Biotech, Shanghai, China) for the genes used in the study. Finally, the experimentally obtained computed tomography (CT) values of lncRNA TEX41, miR-200a-3p, BIRC5 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were analyzed by Graphpad Pism9.5 (GraphPad Software, Santiago, USA) for data analysis using the 2−ΔΔCt normalization method.
Table 1
| Gene | Primer sequence |
|---|---|
| lncRNA TEX41 | Forward: CGTGTCTACACTGGCATGGT |
| Reverse: TCTGGCTATGGGTACTGGA | |
| GAPDH | Forward: CTCTGCTCCTGTTCGAC |
| Reverse: TTCCGTTCTCAGCCTTGAC | |
| MiR-200a-3p | Forward: TAACACTGTCTGGTAACGATGT |
| Reverse: CATCTTACCGGACAGTGCTGGA | |
| BIRC5 | Forward: TTCTGGCTATGTGTGTGTGTGTTCC |
| Reverse: AGTTTGGCTTGCGTCTTCTG |
qPCR, quantitative polymerase chain reaction.
Cell culture and transfection
Human liver cancer cells (SK-Hep-1, LM-3, HepG2, Hep3B, and MHCC-97H) and normal liver cells (QSG-7701) were acquired from Cellverse (Jodhpur, India). The QSG-7701 cells were cultivated in RPMI-1640 (PM150110; Pricella Biotechnology, Wuhan, China) with 10% fetal bovine serum (FBS; 164210; Pricella Biotechnology) and 1% penicillin/streptomycin (PB180120; Pricella Biotechnology). SK-Hep-1, LM-3, HepG2, Hep3B, and MHCC-97H cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; PM150210; Pricella Biotechnology) supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were grown in a 37 ℃ incubator containing a 5% CO2 atmosphere. A TEX41 interference (sh-TEX41) lentiviral vector and corresponding negative control (NC) vector (sh-NC) were provided by Jikai Gene Chemical Technology Co., Ltd. (Shanghai, China). Three small hairpin RNA (shRNA) fragments targeting TEX41 were inserted into hU6-MCS-Ubiquitin-EGFP-IRES-puromycin, and an NC group was used. Sh-TEX41-1 was used in a series of subsequent experiments due to its significant and stable knockdown effect.
Cell Counting Kit-8 (CCK-8) and colony formation detection
In the CCK-8 assay, liver cancer cells SK-HEP-1 and HCCLM3 were cultivated in a 96-well plate (3×102/well; 5 replicates per well) and cultured at 37 ℃. To measure proliferation, a CCK-8 test reagent (C6005; NCM Biotech, Newport, RI, USA) was added to the cells at the same time of day for 3–4 days. A microplate absorbance reader (Bio-Rad Laboratories, Hercules, CA, USA) was used to detect the optical density at 450 nm.
For colony formation assay, liver cancer cell lines SK-HEP-1 and HCCLM3 (1×103 cells/well) were seeded in six-well plates and grown for 14 days. When macroscopic colonies were visible at the bottom of the dish, the experiment was terminated. The cells were fixed with 4% paraformaldehyde for 15 min. Next, 1% crystal violet was added for 15 min of staining. Number of colonies in each was counted, performing these analyses in triplicate.
Wound healing test
HCC cell lines SK-HEP-1 and HCCLM3 (3×105/cell) were seeded into a six-well plate and grown to a confluence of 80–90%. While the cells were being kept in serum-free culture medium, a pipette tip was used to scratch parallel lines in a monolayer. Wound healing was monitored with an inverted microscope on the same day for 3 days. The strength of cell migration ability was determined according to the degree of reduction in wound width and was analyzed via ImageJ software.
Migration and invasion in vitro
For the migration experiment, cells from each group were collected 48 hours after transfection. The cells were resuspended in DMEM without FBS, and the cell density was adjusted to 1×105 cells/mL. A 200-µL cell suspension was inoculated into the upper chamber of a Transwell chamber, and 700 µL of DMEM containing 10% FBS was placed into the lower chamber. The cells were cultured in an incubator with a 5% CO2 atmosphere at 37 ℃ for 48 hours. The cells invading the outer side of the upper ventricular membrane were fixed with 4% paraformaldehyde for 15 min, stained with 0.1% crystal violet, observed under an inverted microscope, and photographed. Five visual fields were randomly selected for cell counting. These procedures were repeated at least three times.
For the invasion experiment, matrix glue and culture medium were mixed at a ratio of 1:8, and diluted substrate gel was added into the upper chamber. After solidification, the cell suspension (1×105 cells/mL; 200 µL) was inoculated in the upper chamber. Subsequently, 500 µL of culture medium containing 10% FBS was added to the lower chamber, and the cells were incubated for 48 hours. We fixed the invading cells by soaking in 4% paraformaldehyde for 15 min. Subsequently, we stained the cells by soaking them in 0.1% crystal violet for 15 min. A microscope was used to count five randomly selected areas to determine the number of invading cells. The experiment was repeated three times.
Western blotting
Proteins were separated using cell protein lysate (P0013B; Beyotime) and protease inhibitor mixture (P1005; Beyotime). After sodium dodecyl sulfate polyacrylamide gel electrophoresis, the protein was transferred to a polyvinylidene fluoride membrane. The membrane was sealed with 5% nonfat milk in Tris-buffered Saline Tween (TBST) for 1 hour and then incubated with antibodies against GAPDH (1:10,000, Proteintech, Rosemont, IL, USA), E-cadherin (1:20,000, Proteintech), N-cadherin (1:2,000, Proteintech) and Vimentin (1:20,000, Proteintech) at 4 ℃ for 2 hours. After four washes with TBST (10 min each time), the proteins were incubated with anti-rabbit (1:10,000, Proteintech) or anti-mouse immunoglobulin G (IgG; 1:10,000, Proteintech) at room temperature for 1.5 hours and subjected to detection staining. The GAPDH protein was used as an internal control.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 8.0 (Dotmatics, Boston, MA, USA). The measurement data of continuous variables are expressed as the mean ± standard deviation. The analysis of continuous variables was conducted via the t-test and repeated-measures analysis of variance. Categorical variables are expressed as numbers and were compared with Chi-squared tests. Survival curves were drawn according to the Kaplan-Meier method, and the log-rank test was used to determine significance, with a P value <0.05 indicating a statistically significant difference.
Results
The expression of lncRNA TEX41 in HCC tissues and cell lines
In order to investigate the differentially expressed genes in liver cancer tissues, we conducted correlation analysis on the dataset GSE41804 in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and found that LINC01093, lncRNA TEX41, LINC00238, LINC00844, and LINC01419 were differentially expressed in HCC. Through PubMed search, it was found that there is currently no research on the role and mechanism of lncRNA TEX41 in HCC. Therefore, this study targeted lncRNA TEX41 to explore its specific molecular regulatory mechanisms in the occurrence, development, and metastasis of liver cancer. We first discovered through the GEPIA2 database (http://gepia2.cancer-pku.cn/#index) that lncRNA TEX41 is significantly overexpressed in HCC tissues (Figure 1A). lncRNA TEX41 was highly expressed in the HCC cell lines, especially in SK-Hep-1 and HCCLM3 (Figure 1B).

The relationship between the relative expression level of lncRNA TEX41 and clinical pathological features in HCC tissues
We collected clinical and pathological information from 50 patients, such as sex, age, tumor size, lymph node metastasis and TNM stage. Through analysis, we found that the high relative expression level of lncRNA TEX41 is significantly correlated with lymph node metastasis and TNM stage in HCC patients. The clinical pathological features of the HCC patients are summarized in Table 2.
Table 2
| Clinicopathological characteristic | Classification | LncRNA TEX41 expression | Total | χ2 | P | |
|---|---|---|---|---|---|---|
| Low (n=16) | High (n=34) | |||||
| Sex | Female | 6 | 5 | 11 | 2.100 | 0.15 |
| Male | 10 | 29 | 39 | |||
| Age (years) | >60 | 11 | 16 | 27 | 2.061 | 0.15 |
| ≤60 | 5 | 18 | 23 | |||
| Tumor size (cm) | <5 | 9 | 28 | 37 | 2.616 | 0.11 |
| ≥5 | 7 | 6 | 13 | |||
| Lymph node metastasis | No | 12 | 15 | 27 | 4.177 | 0.041* |
| Yes | 4 | 19 | 23 | |||
| TNM stage | I & II | 12 | 14 | 26 | 4.987 | 0.02* |
| III & IV | 4 | 20 | 24 | |||
| Total | – | 16 | 34 | 50 | ||
*, P<0.05. TNM, tumor-node-metastasis.
The success of silencing lncRNA TEX41 in SK-HEP-1 and HCCLM3 cells
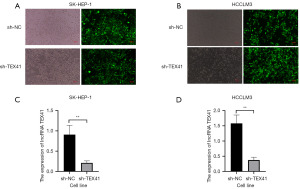
We found that the virus was successfully transfected into SK-Hep-1 and HCCLM3 cell lines via fluorescence microscopy (Figure 2A,2B). The expression level of lncRNA TEX41 in the sh-TEX41 group was lower than that in the sh-NC group (Figure 2C,2D). This also showed that we successfully silenced lncRNA TEX41 by transfecting lentivirus.
The effect of silencing lncRNA TEX41 on the proliferation, migration, and invasion of SK-HEP-1 and HCCLM3 cells
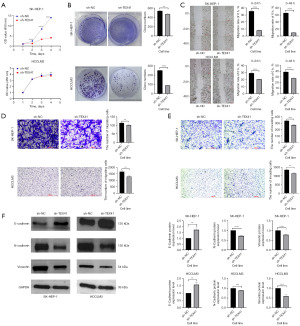
Through CCK8 experiments and cell colony formation experiments, we found that the cell proliferation ability of the sh-TEX41 group was weaker than that of the sh-NC group in both the SK-HEP-1 and HCCLM3 cell lines (Figure 3A,3B). This indicates that silencing lncRNA TEX41 could inhibit the proliferation ability of SK-HEP-1 and HCCLM3 cells. In addition, scratch experiments and Transwell experiments showed that silencing lncRNA TEX41 could inhibit the migration and invasion ability of the SK-HEP-1 and HCCLM3 cell line (Figure 3C-3E). We also examined the effect of TEX41 interference on key proteins involved in epithelial-mesenchymal transition (EMT). As a result, we found that TEX41 knockout increased E-cadherin levels and inhibited N-cadherin and vimentin levels (Figure 3F). This indicates that knockdown of TEX41 can reduce the metastasis of the HCC cell lines SK-HEP-1 and HCCLM3.
Screening and validation of downstream target genes of lncRNA TEX41
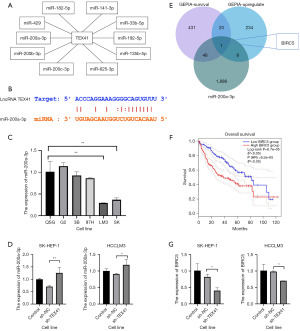
We screened miRNAs that interact with lncRNA TEX41 through the starBase database (Figure 4A), predicted the interaction sites between lncRNA TEX41 and miR-200a-3p (Figure 4B), and found a linear correlation in their expression in multiple HCC samples. Through qRT-PCR experiments, the results showed that miR-200a-3p was significantly downregulated in SK-HEP-1 and HCCLM3 (Figure 4C). After knocking down lncRNA TEX41, miR-200a-3p expression was significantly increased (Figure 4D). We speculated that there may be an interaction between lncRNA TEX41 and miR-200a-3p. Possible downstream target gene BIRC5 was screened using GEPIA2 and starBase databases, it was associated with the overall survival of HCC patients (Figure 4E,4F). qRT-PCR experiments showed that the expression of BIRC5 was significantly reduced after knocking down lncRNA TEX41 (Figure 4G). We speculate that there may be a relationship between lncRNA TEX41 and miR-200a-3p-BIRC5.
Discussion
HCC is among the most common malignant tumors. Despite considerable advance in both basic and clinical in recent years, the incidence and mortality rates of liver cancer have not significantly decreased; in fact, they have risen year on year. Currently, the primary treatment methods for liver cancer include surgery, liver transplantation, radiotherapy, and chemotherapy (15). With the development of detection and treatment methods, the 5-year survival rate of patients with liver cancer is increasing year on year. There is a need to seek new diagnostic biomarkers to assist in early diagnosis and treatment. The human liver cancer cell lines HCCLM3 and SK-HEP-1 are often used to study the molecular mechanisms of HCC; for instance, they are used in studies focused on screening molecular markers for early diagnosis or the identification of targeted therapeutic targets. We aim to provide a new theoretical basis and clinically valuable targets for the diagnosis and treatment of HCC.
LncRNA figures prominently in the occurrence and development of tumors, not only participating in the regulation of epigenetics, cell cycle, and cell differentiation, but also regulating gene transcription and expression in chromatin modification, transcriptional activation, and interference (16). Therefore, lncRNA is considered a relatively novel diagnostic biomarker and therapeutic target for various cancers. With the rapid development of gene chips and next-generation sequencing technology, a growing number of lncRNAs are being identified and studied. In the past few years, a large number of studies have shown that lncRNA is essential to the proliferation, apoptosis, invasion, and metastasis of HCC, providing a new theoretical basis for the diagnosis and treatment of HCC (17-19). LncRNA TEX41 plays a cancer-promoting role in various malignant tumors, but its mechanism of action in HCC remains unclear.
This study analyzed the effect of lncRNA TEX41 on liver cancer cells through cell experiments. The results showed that the low expression of lncRNA TEX41 could inhibit the proliferation, migration, and invasion of the HCCLM3 and SK-HEP-1 HCC cell lines, further confirming the tumor-promoting effect of lncRNA TEX41 in HCC. One study proposed an ceRNA mechanism in which lncRNA and miRNA interact to form a competitive network (20), and this was further confirmed in various subsequent studies. For instance, Yang et al. found that lncRNA TEX41 can upregulate Atg5 through sponging miR-153-3p, thereby promoting the proliferation and migration of squamous cell carcinoma cells (21). In addition, a study has found that lncRNA TEX41 can promote the malignant behavior of melanoma cells by targeting the miR-103a-3p-C1QB axis (22). We examined the miRNAs that interact with lncRNA TEX41 in liver cancer through the use of the starBase database and found that miR-200a-3p plays a role in the occurrence and development of liver cancer. Other research (23) indicates that circBACH1 competes endogenously with miR-200a-3p to promote the replication of hepatitis B virus and the development of liver cancer. Meanwhile, other work (24) has found that miR-200a-3p can be used in the diagnosis and prognostic evaluation of liver cancer and that it is correlated with tumor stage. In our study, we found that silencing lncRNA TEX41 in SK-HEP-1 and HCCLM3 cells resulted in increased expression of miR-200a-3p and inhibited the proliferation and migration of liver cancer cells; this indicates a similar competitive effect between lncRNA TEX41 and miR-200a-3p as that reported previously. In addition, we predicted the target gene BIRC5 of miR-200a-3p using the starBase and GEPIA2 databases. One study reported that BIRC5, as an autophagy-related gene, plays an important role in predicting the prognosis of liver cancer (25). In addition, Li et al. indicated that BIRC5 is a target for the diagnosis and prognosis of hepatitis B virus-related HCC (26). In our study, we found that silencing lncRNA TEX41 reduced the expression of BIRC5 in the SK-HEP-1 and HCCLM3 HCC cell lines.
Conclusions
In summary, silencing lncRNA TEX41 may inhibit the biological functions of liver cancer cells, and its mechanism may involve the targeting of the miR-200a-3p-BIRC5 molecular axis. However, although we found that silencing lncRNA TEX41 may affect the biological behavior of the SK-HEP-1 and HCCLM3 cell lines through the miR-200a3p-BIRC5 axis, this was only examined at the cell level. Therefore, further validation will be conducted at the in vivo level.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-812/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-812/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-812/prf
Funding: This study was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-812/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. This study was approved by the Medical Ethics Committee of The Second Affiliated Hospital of Nantong University (No. 2024KT186). Informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Ajoolabady A, Tang D, Kroemer G, et al. Ferroptosis in hepatocellular carcinoma: mechanisms and targeted therapy. Br J Cancer 2023;128:190-205. [Crossref] [PubMed]
- Aprile M, Costa V, Cimmino A, et al. Emerging role of oncogenic long noncoding RNA as cancer biomarkers. Int J Cancer 2023;152:822-34. [Crossref] [PubMed]
- Mas AM, Huarte M. Long Noncoding RNA Signatures as Cancer Biomarkers. J Clin Oncol 2023;41:3059-62. [Crossref] [PubMed]
- Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov 2021;20:629-51. [Crossref] [PubMed]
- Li W, Qi Y, Cui X, et al. Characteristic of HPV Integration in the Genome and Transcriptome of Cervical Cancer Tissues. Biomed Res Int 2018;2018:6242173. [Crossref] [PubMed]
- Orlandella FM, Smaldone G, Salvatore G, et al. The lncRNA TEX41 is upregulated in pediatric B-Cells Acute Lymphoblastic Leukemia and it is necessary for leukemic cell growth. Biomark Res 2021;9:54. [Crossref] [PubMed]
- Chen ZY, Huang JQ, Zhu Y, et al. Comprehensive Analysis of the Immune Implication of TEX41 in Skin Cutaneous Melanoma. Dis Markers 2021;2021:2409820. [Crossref] [PubMed]
- Jiang Z, Liu H. Metformin inhibits tumorigenesis in HBV-induced hepatocellular carcinoma by suppressing HULC overexpression caused by HBX. J Cell Biochem 2018;119:4482-95. [Crossref] [PubMed]
- Huang N, Fang J, Du F, et al. Uncovering essential anesthetics-induced exosomal miRNAs related to hepatocellular carcinoma progression: a bioinformatic investigation. BMC Med Genomics 2024;17:154. [Crossref] [PubMed]
- Ye HB, Ma BJ, Meng GQ, et al. Bioinformatics analysis of BIRC5 in human cancers. Ann Transl Med 2022;10:888. [Crossref] [PubMed]
- Ren X, Feng N. Unveiling novel prognostic biomarkers and therapeutic targets for HBV-associated hepatocellular carcinoma through integrated bioinformatic analysis. Medicine (Baltimore) 2024;103:e40134. [Crossref] [PubMed]
- Hossen MA, Reza MS, Rana MM, et al. Identification of most representative hub-genes for diagnosis, prognosis, and therapies of hepatocellular carcinoma. Chin Clin Oncol 2024;13:32. [Crossref] [PubMed]
- Li Q, Liu H, Wang H, et al. Anti-BIRC5 autoantibody serves as a valuable biomarker for diagnosing AFP-negative hepatocellular carcinoma. PeerJ 2024;12:e17494. [Crossref] [PubMed]
- Magyar CTJ, O’Kane GM, Aceituno L, et al. Liver Transplantation for Hepatocellular Carcinoma: An Expanding Cornerstone of Care in the Era of Immunotherapy. J Clin Oncol 2025;43:589-604. [Crossref] [PubMed]
- Mattick JS, Amaral PP, Carninci P, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol 2023;24:430-47. [Crossref] [PubMed]
- Chen N, Li Y, Li X. Dynamic role of long noncoding RNA in liver diseases: pathogenesis and diagnostic aspects. Clin Exp Med 2025;25:160. [Crossref] [PubMed]
- Li Y, Shi M, Bie B, et al. NRF1-Induced lncRNA DDX11-AS1 Contributes to the Progression of Hepatocellular Carcinoma via Activating CA9 Expression and the MEK/ERK Pathway. J Hepatocell Carcinoma 2025;12:891-908. [Crossref] [PubMed]
- Su T, Zhang N, Wang T, et al. Super Enhancer-Regulated LncRNA LINC01089 Induces Alternative Splicing of DIAPH3 to Drive Hepatocellular Carcinoma Metastasis. Cancer Res 2023;83:4080-94. [Crossref] [PubMed]
- Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun 2017;8:14421. [Crossref] [PubMed]
- Yang W, Chen Y. LncRNA TEX41 promotes the proliferation and migration of squamous cell carcinoma cells by upregulating Atg5 via sponging miR-153-3p. Cell Mol Biol (Noisy-le-grand) 2023;69:248-54. [Crossref] [PubMed]
- Zheng Y, Zhou W, Li M, et al. IRF4-activated TEX41 promotes the malignant behaviors of melanoma cells by targeting miR-103a-3p/C1QB axis. BMC Cancer 2021;21:1339. [Crossref] [PubMed]
- Du N, Li K, Wang Y, et al. CircRNA circBACH1 facilitates hepatitis B virus replication and hepatoma development by regulating the miR-200a-3p/MAP3K2 axis. Histol Histopathol 2022;37:863-77. [Crossref] [PubMed]
- Yerukala Sathipati S, Aimalla N, Tsai MJ, et al. Prognostic microRNA signature for estimating survival in patients with hepatocellular carcinoma. Carcinogenesis 2023;44:650-61. [Crossref] [PubMed]
- Zhou Y, Shan R, Xie W, et al. Role of autophagy-related genes in liver cancer prognosis. Genomics 2024;116:110852. [Crossref] [PubMed]
- Li S, Hao L, Hu X, et al. A systematic study on the treatment of hepatitis B-related hepatocellular carcinoma with drugs based on bioinformatics and key target reverse network pharmacology and experimental verification. Infect Agent Cancer 2023;18:41. [Crossref] [PubMed]
(English Language Editor: J. Gray)


