
The p53 family in hepatocellular carcinoma
Hepatocellular carcinoma (HCC): epidemiology, risk factors and current treatment strategies
HCC is the sixth most common cancer globally and the third leading cause of cancer-related death worldwide with a steadily increasing annual incidence of about 900,000 cases per year (1). HCC is the deadly complication of chronic liver disease and usually develops within liver cirrhosis related to different etiologies. In Asia and Africa hepatitis B virus (HBV) infection, with or without aflatoxin B1 exposure, is the most frequent etiology, whereas hepatitis C virus (HCV) infection, chronic alcohol abuse and metabolic syndrome are most frequently related to HCC in Western countries (1-4).
Because more than 90% of cancers arise in patients with underlying cirrhosis, treatment of HCC requires both the management of the malignant liver tumor and the underlying liver disease. Consequently, the selection of treatment modalities depends not only on the stage of tumor and the performance status of the patient but also on the underlying liver function. The Barcelona Clinic Liver Cancer (BCLC) classification stratifies patients accordingly and provides therapeutic algorithms (5). Surgery, liver transplantation and ablative strategies are the mainstays of a curative approach for early HCCs (6). Transarterial chemoembolization (TACE) can be offered to patients with intermediate stage HCC no longer amenable to cure (6). Up to date, the tyrosine kinase inhibitor sorafenib is the only available standard of care for systemic treatment of advanced HCC. An effect of sorafenib in an adjuvant setting could not be confirmed. Data of a phase II trial that has been recently published as abstract confirm the efficacy of regorafenib as second line treatment (Phase III RESORCE Trial). HCC is in need for additional molecular treatments in first- and second-line therapy and also in the adjuvant setting. New data from international multi-centric genome sequencing projects are suggesting further promising therapeutic targets. For individualized patient care, genomic alterations identified in targetable genes will be useful to identify patients with HCC who could benefit from specific targeted therapies in clinical trials.
Data from high-throughput analyses have provided us with an understanding of the genetic landscape of HCC genetic alterations and the key events that regulate tumor development, progression and treatment response. Translation of this knowledge into novel druggable targets, new biomarkers and medical decision making might ultimately improve patient care and survival.
Most frequent mutations in HCC affect TERT promoter (60%) associated with an increased telomerase expression. TP53 and CTNNB1 are the next most prevalent mutations affecting 25–30% of HCC patients. Low frequency-mutated genes include e.g., AXIN1, ARID2, ARID1A, TSC1/TSC2, RPS6KA3, KEAP1, and MLL2. Association of mutations helped define three groups of genes related to risk factors and centered on CTNNB1 (alcohol), TP53 (HBV) and AXIN1 (7).
Recent whole-exome sequencing studies allowed the identification of the major pathways mutated in HCC. These are (I) telomere maintenance; (II) Wnt/β-catenin pathway; (III) p53 cell cycle pathway; (IV) epigenetic modifiers; (V) oxidative stress pathway; (VI) PI3-K/AKT/MTOR and RAS/RAF mitogen-activated protein kinase pathways (8). In the following we will focus on the p53 pathway.
The p53 cell cycle pathway is altered in at least half of HCC patients. Disruption of the p53 pathway in HCC can either occur by mutations of the p53 gene itself (9-11) or by alterations such as p14 ARF inactivation (12) or as a result from the amplification/overexpression of its specific inhibitors MDM2 and MDM4 (13,14).
The p53 family: isoforms, functions and regulation
The importance of p53 is indicated by the fact that p53 is the most frequently mutated gene in human cancer. In addition to the 50% of cases where p53 is mutated, there are 20% of cancer cases where p53 is functionally inactivated. At the protein level, p53 and its family member p63 and p73 act as transcription factors. p53 protein structure, transcriptional targets and mechanisms of binding and activating different promoters have been intensely studied. Tumor suppressor protein p53 was originally described in 1979 and since then its function has expanded from the “guardian of the genome” to regulation of a variety of cellular processes, including metabolism, cell cycle, cell death, senescence, and the production of cytokines and inflammatory proteins (15-18). Thus, p53 not only acts as a tumor suppressor but also influences development, neurodegeneration, aging, autophagy, angiogenesis, maternal reproduction and fertility among other physiological and pathophysiological conditions. p53 controls these essential processes primarily based on its function as a transcriptional regulator of an expanding series of downstream target genes. Since the description of the p53 DNA-binding consensus element (RRRCWWGYYY) (19) and the first identified p53 responsive gene p21CIP1 (CDKN1A) (20), which plays a role in inducing G1 cell cycle arrest upon DNA damage, approximately 150 genes have been reported to be regulated by p53. In addition to p21CIP1, relevant genes controlling cell cycle, cell death and apoptosis have been validated. These include Mdm2 (the essential negative regulator of p53), CD95 and PUMA (21-24).
There are now more than 28,000 TP53 mutations that have been described in human cancer (IARC TP53 Database) (25). Mutations in the p53 gene may lead to three types of cellular outcome. (I) The p53 mutation can result in loss of wildtype activities, i.e., activation of p53 target genes and loss of tumor suppressor function; (II) many mutant p53 isoforms can exert dominant-negative effects over wild type p53 expressed from the remaining wildtype allele; (III) some mutant p53 isoforms carry new pro-oncogenic activities referred to as “gain-of-function” (GOF). These GOF properties are not shared by wildtype p53, and are independent of their ability to exert a dominant-negative effect on wildtype p53.
In 1997, the two “younger siblings” of p53, p63 and p73 were identified, displaying high homology to p53, for which reason these three proteins were defined to form the p53 family. The discovery that the p53 family consists of three members (TP53, TP63 and TP73) increased the complexity of this network, as the two p53 homologs also contribute to the tumor suppressor and the oncogenic potential of p53Jeny (26-28). Both, TP63 and TP73 are expressed as many isoforms due to alternative usage of promoters for transcription and alternative splicing. Long isoforms of p63 and p73 containing a transactivation domain (TAD) (TAp63 or TAp73) are able to transactivate the same target genes as p53 and induce apoptosis. In contrast, short, amino-terminally deleted dominant negative p63 or p73 isoforms (DNp63 or DNp73) have opposing effects via DN mechanisms. p53 family proteins share the modular composition of a DNA binding domain (DBD), a proline-rich domain (PRD), an oligomerization domain (OD), and a TAD. Additionally, a transactivation inhibitory domain (TID) and a sterile alpha motif (SAM) domain were described for some isoforms of p63 and p73, while a basic region (BR) is present in individual p53 isoforms (29). The DBD represents the essential part for the transcriptional activity of the proteins. Regulatory binding sites are located within the TAD, whereas splicing and post-translational modifications preferentially occur within the OD of p53 family members (29). SAM and TID domains are important for activity regulation by influencing protein formation (30) (Figure 1).
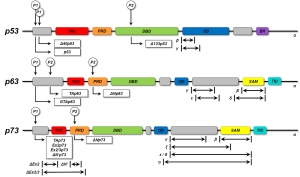
By using multiple promoters and alternative splicing p53 family genes produce a diversity of isoforms. In summary, full-length isoforms contain a functional TAD and are capable to activate downstream target genes and induce apoptosis. TP63 and TP73 cooperate with TP53 to induce apoptosis, generating a complex network of interactions between the products of these three genes. In contrast, amino-terminally truncated DN isoforms of p63 and p73 are potent inhibitors of the full-length p53 family isoforms by preventing their transactivation activity and thus the induction of apoptosis (31). An additional layer of complexity is added by the diversity of p53 mutants. Among these, p53 mutants with unfolded structure, but not DNA contact mutants, bind specifically to p63 and p73 and thus abrogate their apoptotic activity.
p53 in HCC
In search for novel therapeutic approaches, the molecular knowledge of hepatocarcinogenic events is increasingly translated into clinical trials. p53 is mutated in more than 50% of aflatoxin B1-induced HCC, in up to 45% of HBV-related HCC and in 13% of HCV-related HCC (2). Preferential mutation sites are located within the DBD of p53, reducing its binding affinity to responsive elements and therefore leading to decreased expression of p53 target genes (32).
We have previously shown that the p53 family members p63 and p73 do not only play an important role in embryonic development and differentiation but also in induction of apoptosis and treatment response of HCC (31,33,34). Activation of the p53 family is a central event in the DNA-damage response, chemosensitivity and prognosis of HCC. p53 family-mediated apoptosis signaling is affected at multiple levels in HCC. Mutation of p53 and an altered ratio of TA versus DN-isoforms of p63 and p73 confer therapy resistance and lead to poor prognosis of patients with HCC (35-37). Interference of DNp73 with apoptosis-/chemosensitivity takes place at several levels of apoptosis signaling. DNp73 negatively regulates the genes encoding for the death receptors CD95, TNF-R1, TRAIL-R2 and TNFRSF18 (38). Furthermore, DNp73 represses the genes encoding caspase-2, -3, -6, -8 and -9 (38). Concomitantly, DNp73β inhibits apoptosis emanating from mitochondria (38). Thus, DNp73 expression in HCC selects against both the death receptor and the mitochondrial apoptosis activity of the TA isoforms. The clinical importance of these data is evidenced by our finding that the DNp73β target gene signature can predict the prognosis of patients suffering from HCC (38). Furthermore and of clinical relevance, essential mechanism of mutant p53 GOF activity in HCC is the ability of these mutants to bind and inactivate the TA isoforms of p63 and p73 (28,39). Thus, p53 GOF mutants affect the p53 family-mediated regulation of pro-apoptotic genes controlling the extrinsic and intrinsic apoptosis signaling pathways in HCC contributing to impaired apoptosis and drug resistance. Therefore, targeting the interaction of GOF mutant p53 proteins with TAp63 and TAp73 seems a promising strategy for future HCC therapy.
p53 family as a target for novel therapeutic strategies in HCC
Targeting wildtype p53
A crucial goal in the development of anticancer strategies in HCC must therefore be the restoration of physiologic apoptosis in response to cellular stress signals by reconstitution of the tumor suppressor function of p53 family members. In cancer cells harboring wildtype p53, this might succeed with the “simple” recovery of wildtype p53 function. In a recent study, TACE combined with the injection of recombinant adenovirus p53 was described as an effective therapy in patients with unresectable HCC, contributing to improved overall survival and progression-free survival rates compared to TACE monotherapy (40). Furthermore, a number of small molecules have been identified, which are able to restore wildtype p53 function to cancer cells (Figure 2).
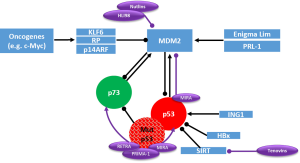
A key target for these approaches is the p53/MDM2 interface. The first small molecule inhibitors which target the p53/MDM2-interaction are Nutlins. Nutlins are a family of three (Nutlin-1, Nutlin-2, Nutlin-3) cis-imidazoline analogs. They occupy the essential hydrophobic pocket of MDM2 that mediates p53 interaction (41). Thus, Nutlins prevent p53 degradation and lead to p53 stabilization and accumulation. Interestingly, apart from p53 induction, Nutlin-3 is also capable to induce apoptosis in HCC cells by direct activation of p73 via disruption of p73-MDM2 binding (16,42).
Another small molecule targeting the p53/MDM-interaction is RITA (reactivation of p53 and induction of tumor cell apoptosis). RITA binds directly to the N-terminus of p53 and thereby induces conformational changes, leading to an abrogation of MDM2 association (16,31). The spiro-oxindole MI-219 is another highly specific small molecule inhibitor of p53/MDM2-interaction and additionally induces auto-ubiquitination and proteasomal degradation of MDM2 (16,43).
As an alternative to interfering with p53/MDM2-interaction HLI98 (HDM2 ligase inhibitor 98) molecules and their derivatives named MDP compounds bind to the C-terminal RING-domain of MDM2 and inhibit its E2 ligase activity. Thus, p53 ubiquitination and degradation is prevented (16). Attention should be paid to the fact that stabilization and activation of p53 also results in the induction of the auto-regulatory feedback-loop, leading to increased production of MDM2. So far, no data are available concerning the effect on re-produced MDM2 and therefore the long term efficacy of these drugs.
A new strategy to restore p53 function independent of MDM2 is the blockade of SIRT1, a nicotinamide adenine dinucleotide-dependent class III histone deacetylase, which inactivates p53 by deacetylation. Two small molecules, Tenovin-1 and -6, have been shown to inhibit SIRT1 function and to induce apoptosis (16).
Targeting mutant p53
Pharmacological reactivation of mutant p53 in cancer is a clear challenge. Mutant p53 is a different kind of target compared with other successful novel anticancer therapies such as Herceptin (trastuzumab) and Gleevec (imatinib) which block oncogenic kinases overexpressed in different tumors. However, for mutant p53, the issue is to refold and reactivate a nonfunctional tumor suppressor (44). Furthermore, this target is a DNA-binding transcription factor that has been considered not easily druggable. However, the fact that mutant p53 can exert GOF functions opens up the perspective to inhibit these functions. This may be easier to accomplish than restoration of wildtype function.
Two small molecules—PRIMA-1 (p53 reactivation and induction of massive apoptosis) and MIRA-1 (mutant p53 reactivation and induction of rapid apoptosis)—are capable to restore and stabilize the original DBD, refold mutant p53 and enhance expression of several TP53 targets, including Bax, PUMA and Noxa. In HCC cell line cells expressing mutant p53 cytotoxic effects of PRIMA-1 were described, however without a restoration of wildtype DNA binding and transcriptional activities (45). MIRA-1 is structurally not related to PRIMA-1, but shares the capacity to reconstitute DNA binding and transcriptional activities of p53 (16). RETRA (reactivation of transcriptional reporter activity), originally designed to re-establish p53 function, was shown to elevate TAp73 levels by dissociation of an inhibitory TAp73/mutant p53 complex (16). Disruption of complexes of GOF mutant p53 and TP63/p73 by RETRA thus represents another promising approach to restore expression of TP53763/73 target genes and tumor suppression.
Conclusions
The p53 family plays a central role in tumorigenesis, treatment response and prognosis of HCC. Whereas p53 is often mutated in HCC, p63 and p73 function is preserved, yet the effects, tumor-suppressive vs. oncogenic are determined by the ratio and different expression patterns of their TA and DN isoforms. The molecular mechanisms regulating the interplay between the different isoforms of the members of the p53 family and the plethora of p53 mutants are in the focus of current research. In recent years it has been revealed that all members of the p53 family are expressed as a diverse variety of isoforms. Depending on the isoform expressed, the role of a gene can drastically change from a tumor suppressor to an oncogene. Consequently, special emphasis should be put to the DN isoforms of p63 and p73, which have been shown to be of critical importance for carcinogenesis and chemoresistance. Thus, targeting specific p53 family isoforms might be the key to novel therapeutic strategies for HCC and other human cancers.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Zhi-Min Yuan) for the series “p53 Biology and Cancer” published in Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.11.79). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Shiraha H, Yamamoto K, Namba M. Human hepatocyte carcinogenesis Int J Oncol 2013;42:1133-8. (review). [PubMed]
- Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer 2015;15:653-67. [Crossref] [PubMed]
- Kew MC. Hepatocellular carcinoma: epidemiology and risk factors. J Hepatocell Carcinoma 2014;1:115-25. [Crossref] [PubMed]
- Llovet JM, Villanueva A, Lachenmayer A, et al. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015;12:408-24. [Crossref] [PubMed]
- Knox JJ, Cleary SP, Dawson LA. Localized and systemic approaches to treating hepatocellular carcinoma. J Clin Oncol 2015;33:1835-44. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015;149:1226-1239.e4. [Crossref] [PubMed]
- Hussain SP, Schwank J, Staib F, et al. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene 2007;26:2166-76. [Crossref] [PubMed]
- Zalcenstein A, Stambolsky P, Weisz L, et al. Mutant p53 gain of function: repression of CD95(Fas/APO-1) gene expression by tumor-associated p53 mutants. Oncogene 2003;22:5667-76. [Crossref] [PubMed]
- Schilling T, Kairat A, Melino G, et al. Interference with the p53 family network contributes to the gain of oncogenic function of mutant p53 in hepatocellular carcinoma. Biochem Biophys Res Commun 2010;394:817-23. [Crossref] [PubMed]
- Anzola M, Cuevas N, López-Martínez M, et al. P14ARF gene alterations in human hepatocellular carcinoma. Eur J Gastroenterol Hepatol 2004;16:19-26. [Crossref] [PubMed]
- Biderman L, Manley JL, Prives C. Mdm2 and MdmX as Regulators of Gene Expression. Genes Cancer 2012;3:264-73. [Crossref] [PubMed]
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013;13:83-96. [Crossref] [PubMed]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009;9:749-58. [Crossref] [PubMed]
- Pflaum J, Schlosser S, Müller M. p53 Family and Cellular Stress Responses in Cancer. Front Oncol 2014;4:285. [Crossref] [PubMed]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979;278:261-3. [Crossref] [PubMed]
- Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 1979;17:43-52. [Crossref] [PubMed]
- el-Deiry WS, Kern SE, Pietenpol JA, et al. Definition of a consensus binding site for p53. Nat Genet 1992;1:45-9. [Crossref] [PubMed]
- el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993;75:817-25. [Crossref] [PubMed]
- Momand J, Zambetti GP, Olson DC, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992;69:1237-45. [Crossref] [PubMed]
- Jeffers JR, Parganas E, Lee Y, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 2003;4:321-8. [Crossref] [PubMed]
- Müller M, Strand S, Hug H, et al. Drug-induced apoptosis in hepatoma cells is mediated by the CD95 (APO-1/Fas) receptor/ligand system and involves activation of wild-type p53. J Clin Invest 1997;99:403-13. [Crossref] [PubMed]
- Müller M, Wilder S, Bannasch D, et al. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med 1998;188:2033-45. [Crossref] [PubMed]
- Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007;28:622-9. [Crossref] [PubMed]
- Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998;2:305-16. [Crossref] [PubMed]
- Kaghad M, Bonnet H, Yang A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997;90:809-19. [Crossref] [PubMed]
- Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ 2015;22:1239-49. [Crossref] [PubMed]
- Ferraiuolo M, Di Agostino S, Blandino G, et al. Oncogenic Intra-p53 Family Member Interactions in Human Cancers. Front Oncol 2016;6:77. [Crossref] [PubMed]
- Candi E, Agostini M, Melino G, et al. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum Mutat 2014;35:702-14. [Crossref] [PubMed]
- Müller M, Schleithoff ES, Stremmel W, et al. One, two, three--p53, p63, p73 and chemosensitivity. Drug Resist Updat 2006;9:288-306. [Crossref] [PubMed]
- Meng X, Franklin DA, Dong J, et al. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res 2014;74:7161-7. [Crossref] [PubMed]
- Mundt HM, Stremmel W, Melino G, et al. Dominant negative (DeltaN) p63alpha induces drug resistance in hepatocellular carcinoma by interference with apoptosis signaling pathways. Biochem Biophys Res Commun 2010;396:335-41. [Crossref] [PubMed]
- Müller M, Schilling T, Sayan AE, et al. TAp73/Delta Np73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ 2005;12:1564-77. [Crossref] [PubMed]
- Stiewe T, Tuve S, Peter M, et al. Quantitative TP73 transcript analysis in hepatocellular carcinomas. Clin Cancer Res 2004;10:626-33. [Crossref] [PubMed]
- Tannapfel A, John K, Mise N, et al. Autonomous growth and hepatocarcinogenesis in transgenic mice expressing the p53 family inhibitor DNp73. Carcinogenesis 2008;29:211-8. [Crossref] [PubMed]
- Wei J, Zaika E, Zaika A. p53 Family: Role of Protein Isoforms in Human Cancer. J Nucleic Acids 2012;2012:687359.
- Bantel H, Simon HU. DeltaNp73beta is oncogenic in hepatocellular carcinoma by blocking apoptosis signaling via death receptors and mitochondria. Cell Cycle 2010;9:2710-1. [Crossref] [PubMed]
- Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010;2:a001107 [Crossref] [PubMed]
- Shen A, Liu S, Yu W, et al. p53 gene therapy-based transarterial chemoembolization for unresectable hepatocellular carcinoma: A prospective cohort study. J Gastroenterol Hepatol 2015;30:1651-6. [Crossref] [PubMed]
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004;303:844-8. [Crossref] [PubMed]
- Wu H, Leng RP. MDM2 mediates p73 ubiquitination: a new molecular mechanism for suppression of p73 function. Oncotarget 2015;6:21479-92. [Crossref] [PubMed]
- Shangary S, Qin D, McEachern D, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A 2008;105:3933-8. [Crossref] [PubMed]
- Joerger AC, Fersht AR. The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol 2010;2:a000919 [Crossref] [PubMed]
- Shi H, Lambert JM, Hautefeuille A, et al. In vitro and in vivo cytotoxic effects of PRIMA-1 on hepatocellular carcinoma cells expressing mutant p53ser249. Carcinogenesis 2008;29:1428-34. [Crossref] [PubMed]


