
DNA hypermethylation of tumor suppressor genes as an early lung cancer biomarker
Introduction
Despite the advances in lung cancer therapeutics perspectives involving personalized therapies and investigation of cancer biomarkers the available epidemiological data presents a disquieting scenario of the lung cancer mortality and morbidity ratio in the near future. Alarming epidemiological reports concerning lung tumors incidence are probably associated with a still low social awareness of adverse effects of cigarette consumption on a human body and low level of interest in tobacco prevention programs. The major problem is also lack of screening tests which could improve detection of tumor developing in an early stage, what seems to be associated with difficulties in selection of cancer high risk individuals. Other emerging issues noted especially in developing countries are both too high cost of screening programs and still restricted accessibility to advanced imaging diagnostics including computed tomography (CT) or low-dose spiral computed tomography (LDSCT) (1-3).
The mentioned issues influence a late detection of most patients when disease is diagnosed in a locally advanced or advanced stage with presence of distant metastases. Unfortunately tumor detected in a late stage of the disease disqualifies patients from radical surgery and in consequence prevents from complete recovery. In the late-tumor stage patients the treatment options include characterized by restricted efficacy: chemotherapy, radiotherapy or radiochemotherapy (4). However, standard chemotherapy regimen is constantly used routinely in most advanced cases, because the number of patients who could achieve from personalized therapy is still limited (5).
Although in some developed countries the CT/LDSCT-based diagnostics was applied for early lung cancer detection, there is still lack of indications or recommendations to introduce such methods to cancer screening in a general population, though tumor development is often asymptomatic, insidious and also not in each case strongly associated with cigarette consumption. It is worth noting that CT/LDSCT scans may visualize small pulmonary nodules which are usually confirmed as non-cancerous lesions, what may leading to generation of false positive results. The above presumptions may be probably clarified by expanding of diagnostic perspectives with introduction of molecular markers which could provide early detection of lung cancer, reduce false results rate obtained by imaging diagnostics or even be applied as independent diagnostics tools (6,7).
Among potential lung cancer epigenetic biomarkers, hypermethylation of tumor suppressor genes promoters is meticulously investigated in the last decade. Involvement of DNA methylation phenomena in regulation of key processes of the cell cycle allows considering it as potential diagnostic markers of neoplasms including lung cancer. Moreover, the possibility of investigation of hypermethylation in circulating cell-free DNA (cfDNA) using liquid biopsy make it a promising biomarker of lung cancer (8,9).
The role of DNA methylation in cancer development
DNA methylation of gene promoter is a dominant and the most known epigenetic alteration of human genome. The sequences in genome that undergo methylation are not accidental and especially concern cytosines located within CpG islands [repetitive dinucleotide sequences (5'-CG-3')]. It is estimated that approximately half of genome sequences consist of CpGs out of which about 70–80% are methylated. In physiology, activity of DNA methyltransferases (DNMTs) provides a stable methylation pattern in cell genome and thereby controls gene expression post-replication. Consequently the methylation of CpG islands is mostly observed in genes encoding tissue-specific proteins, except from the cells which a gene product is typical to. The genetic information concerning gene expression after cell division (DNA methylation pattern) is inherited by progeny cells. It seems to be a main warranty of gene expression pattern in different tissues—DNA methylation pattern is typical for the defined types of cells. However, the 5’ regions of tumor suppressor genes promoters which are rich in CpGs sequences do not undergo methylation due to their key function in healthy cells cycle. Unmethylation of gene promoter regions is a leading requirement for an active and controlled gene transcription (10-12).
The methylation of promoter region causes gene silencing, what leads to changes in structure of chromatin and its conversion into a condensed and inactive form (heterochromatin). It causes blockade of the promoter transcription start site (TSS), so its recognition by transcription factors and transcription of genetic information from DNA to mRNA cannot be processed. Alterations of a methylation pattern which are finding in tumor cells may result in genetic repression of information encoded in the DNA. However, the methylation pattern of CpGs of tumor cells is variable compared to healthy cells throughout the hypomethylation or hypermethylation which in result determine new DNA methylation pattern. The hypermethylation of tumor suppressor genes promoters seems to be a significant epigenetic alteration noted in tumor cells. As regards to methylation of gene CpGs promoter regions in physiology, the normal cells do not undergo such epigenetic modification because of their crucial function as control mechanisms of cell cycle. Furthermore, protective mechanisms against promoter hypermethylation involving regulation of replication, chromatin modification, demethylation of DNA are demonstrated by non-tumor cells. Mentioned mechanisms efficiently prevent the access of DNMTs to DNA. On the other hand, protective mechanisms against methylation are disabled in tumor cells, what causes circumvent of protective systems by DNMTs. Moreover, the overexpression of DNMTs which are responsible for de novo methylation of promoter CpGs is commonly noted in tumor cells (10-13). Due to a large interest in DNA methylation concerning its involvement in cell cycle and observed disorders in methylation processes in tumor cells, this pre-transcriptional gene modification is currently carefully considered as a potential marker for early lung cancer diagnosis.
Development of DNA hypermethylation as a lung cancer biomarker
Currently imaging diagnostics and patients’ clinical factors are often insufficient for early diagnosis of lung cancer. Unfortunately most of cancer cases are diagnosed in a late stage of the disease and require invasive diagnostics tools (e.g., bronchoscopy with EBUS-TBNA or transthoracic biopsy) to obtain tumor sample (cells or tissue) for assessment of lung cancer histology and conduction of molecular examination. Similarly, invasive diagnostic procedures refer to assessment of histology of undefined small pulmonary nodules detected in a LDSCT/CT screening. Moreover, mentioned invasive diagnostics methods may expose lung cancer patients on periprocedural complications and even revision of minor-surgery in view of the risk of obtaining a non-diagnostic material (lack of tumor cells in the examined sample or degradation of clinical material). It is worth nothing that collected clinical samples are subsequently fixed before examination and then embedded in paraffin blocks or cell-blocks, what may lead to degradation of tumor cells and consequently falsify the results of molecular testing (resulting in manifestation of false positive or false negative results). Based on the above facts clinical material scheduled to molecular testing is preferred to collection with non-invasive manner and should be easy to obtain (14,15).
Although sputum is the easiest diagnostic material to obtain especially in patients with tumor located centrally in mediastinum, its sensitivity in lung cancer diagnosis ranges 22–98% and depends on tumor size, patient’s ability to expectorate sputum and an experience of a pathomorphologist examining specimens (16). Moreover, the sputum is frequently scant in tumor cells what prevents from reliable molecular testing and high specific examination of genes methylation status. In consequence molecular examination of sputum ceased to be a diagnostic standard in early lung cancer detection. Recent studies analyzed hypermethylation status of selected tumor suppressor genes are consistent with above findings. The following genes examined in sputum samples demonstrated the sensitivity and specificity: APC (23.1%/96%), CDH13 (27.6%/75%), CDKN2A (p16) (39.8%/72.8%), DAPK (47.2%/82.2%), MGMT (35.8%/85.6%), RASSF1A (12.2%/93.5%), TCF21 (53.8%/100%), respectively (17-19). However, some studies presented acceptable diagnostic accuracy for assessment of risk of lung cancer development, when methylation was examined simultaneously in loci of a few genes. Belinsky et al. achieved sensitivity and specificity of 64% for lung cancer prediction analyzing methylation status of six following genes p16, MGMT, DAPK, RASSF1A, PAX5β and GATA5. Hypermethylation of mentioned genes was associated with a >50% increased lung cancer risk (18). In another study, designed methylation panel analyzed methylation profile of four genes: 3-OST-2, RASSF1A, p16 and APC in sputum samples allowed to distinguish lung cancer patients from healthy individuals with sensitivity of 62% and specificity of 100% (20).
Currently the most promising seems to be investigation of tumor suppressor genes methylation status using liquid biopsy technique (examination of blood sample) which could provide non-invasive diagnosis of cancer. In contrast to physiology, tumor cells demonstrate an increased cellular metabolism, likewise cells uncontrolled proliferation and partial destruction by immunological mechanisms lead to their disintegration in necrosis and apoptosis mechanism. Therefore significantly higher cfDNA concentration is observed in circulation of cancer patients compared with healthy individuals. In cancer patients the cfDNA concentration positively correlates with tumor stage, size, aggressiveness and presence of distant metastases. Moreover, molecular status of cfDNA with estimated presence of epigenetic alterations reflects molecular status in tumor tissue. High vascularization of the lung tissue and tumor potency to angiogenesis stimulates formation of blood vessels network, which promotes a release of high yield tumor cfDNA into the circulation. The investigation of tumor suppressor genes hypermethylation using liquid biopsy technique is enabled by a high stability of cytosines within cfDNA sequence, and lack of cfDNA hypermethylation in the blood of healthy people (21-23). The recent large studies evaluating methylation status of selected genes as early and non-invasive lung cancer markers are presented in Table 1.
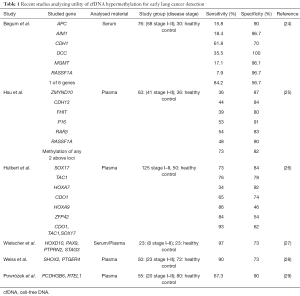
Full table
Hypermethylation of tumor suppressor genes promoter in lung cancer screening
In recent years a few randomized clinical trials were conducted to evaluate utility of imaging diagnostics (RTG) or imaging diagnostics supported by a sputum examination in high risk individuals of lung cancer development. However, that screening improved the detection rate of lung tumors in the stage I of the disease and increased the 5-year survival rate, the decrease in mortality rate of screened individuals was not achieved (16,30). Recent studies proved that more accurate diagnosis of lung cancer can ensure screening by LDSCT. In a randomized trial conducted by the National Lung Cancer Screening Trial (NLST), the mortality reduction of 20% was achieved in a group of patients screened by LDSCT compared to standard chest RTG. According to the NLST, the chances of developing lung cancer with a positive CT result are below 5%, because lung cancer with CT screening in the NLST study achieved 71% sensitivity and 63% specificity with a 96.4% false positive rate (31,32). In up to 50% of individuals screened by the LDSCT small pulmonary lesions (which diameter do not exceed of 10 mm) with a benign etiology are detected. Unfortunately about 20% of these nodules which are scheduled to thoracotomy do not confirm their malignancy, thus LDSCT may lead to false positive results and unnecessary surgery (31-33). In such cases the molecular examination analyzing epigenetic markers could support LDSCT and reduce rate of false results.
To date there is a lack of established recommendations concerning the application of gene hypermethylation in lung cancer screening, but the available literature data seem to confirm their high applicability in a daily clinical practice. Current studies draw the particular attention to the application of epigenetic biomarkers as complementary tests for early imaging diagnostics of lung cancer. The main purpose of the simultaneous application of both methods constitutes an improvement of the sensitivity and specificity of screening and the reduction of false positive/negative results rate. Then, a positive result obtained in a high risk individuals based on the analysis of blood cfDNA methylation may be a first indication to schedule such individuals to the imaging diagnostics. The leading advantage of epigenetic screening over the other examinations is a possibility of their non-invasive detection using a liquid biopsy technique (26-28).
Majority of available papers focused especially on the designation of hypermethylated gene signature which could distinguish cancer patients from healthy individuals or patients with benign lung diseases. Nowadays, only two large studies detailed analyze utility of DNA hypermethylation in lung cancer screening. The NELSON LDSCT trial screened methylation status of sputum DNA of asymptomatic high-risk individuals to detect lung cancer at preclinical stage in a screening interval of 2 years. The selected diagnostic panel of three genes including: RASSF1A, 3OST2 and PRDM14 detected 28% of lung cancer cases within 2 years with specificity of 90%. Sputum cytology examination in contrast to epigenetic screening did not detect any lung cancer cases. As a previously mentioned DNA hypermethylation analysis in sputum may play a potential role in the detection of preclinical disease, but complementary diagnostic markers are needed to improve the low sensitivity (34). The latest study of Hulbert et al. evaluated the utility of plasma and sputum DNA hypermethylation panel as an adjunct test to lung cancer CT screening. Interestingly, the sputum diagnostic panel of three following genes: TAC1, HOXA7 and SOX 17 demonstrated a high diagnostic accuracy for early lung cancer detection (stage I–II of the disease) with sensitivity of 98% and specificity of 71% (AUC =0.890) with high negative predictive value (NPV) and positive predictive value (PPV) of 89% and 93%, respectively. Additionally, the plasma cfDNA methylation panel including genes: TAC1, CDO1 and SOX17 presented following diagnostic accuracy for early lung cancer: 93% sensitivity, 62% specificity (AUC =0.770), NPV and PPV 78% and 86%, respectively. Moreover, independent blinded random prediction model combining gene methylation of above genes with clinical factors correctly predicted lung cancer in 91% of individuals using sputum detection and 85% of individuals using plasma detection. Cited study confirmed that, designed methylation panels could be used as a complementary to CT screening, identifying individuals at high risk for lung cancer, reducing false positive results, unnecessary toracotomies and improving the diagnosis of tumor at an earlier stage (26).
The above mentioned data seem to confirm the importance of epigenetic tests in early lung cancer detection and their applicability in screening programs. Careful designation of diagnostic tests may significantly contribute to an improvement of lung cancer detection statistics and lead to a reduction of false results obtained by LDSCT.
Current status and future perspectives
To date hypermethylation of various tumor suppressor genes was investigated as potential lung cancer biomarkers. High methylation frequency of e.g., CDKN2A (p16), MGMT, DCLK1, CDH13, RASSF1A, RARB2 and many others was noted (35-40). Despite the potential utility of above genes in lung tumors detection, these are also widely hypermethylated in other cancers. Therefore the leading challenge in application of methylated genes into routine diagnostics is a determination of tumor specific genes, which undergo hypermethylation only in selected tissue-specific tumors. In consequence, it is difficult to assign hypermethylation of particular genes to defined disease, what is a main limitation of use these in daily clinical practice. An ideal example for that issue seems to be hypermethylation of SEPT9 which is an established epigenetic marker of colorectal cancer and used in its diagnostics. However, recently hypermethylation of this gene was found in lung cancer patients, what undermines its colorectal cancer specificity and put a red flag for diagnostic tests assessing methylation status of SEPT9 (41). The next issue is a total number of CpG islands which could be methylated and their location within promoter sequences. Firstly methylation pattern may depend on cell type which is from cancer development. Secondly methylation process may be restricted to selected CpGs in different cancers. Moreover, the methylation signature may differ between tumor tissue and blood cfDNA (42). The tool which could advance application of cfDNA methylation analysis in daily practice is a wide genome methylation sequencing of DNA and its comparison between tissue and blood samples of cancer patients and healthy individuals. Perhaps, such procedure could provide novel findings which will lead to designation of diagnostic epigenetic based tests.
The next challenge is a validation of previously selected biomarkers into clinics. As noted above process of biomarkers selection for lung cancer detection will required long-drawn and laborious validation process conducted in a large group of lung cancer patients and healthy individuals and even patients suffered from other cancers. Moreover, biomarkers validation should be conducted by a large independent diagnostic center with application of complicated diagnostic methods such as DNA microarrays and next-generation sequencing. Such considerations seem to be confirmed by a fact that generally only two epigenetic diagnostics tests were registered for early diagnosis or confirmation of tumor presence based on molecular alterations analysis. Mentioned presence of SEPT9 methylation in cfDNA was established as a diagnostic marker of colorectal cancer, whereas the analysis of short stature homeobox 2 gene (SHOX2) methylation in bronchoalveolar lavage samples may be a confirmatory test of ambiguous result of cytology examination for lung cancer detection. Although both tests were certified to in vitro diagnostics (CE-IVD certificate), their diagnostic accuracy is limited. Unfortunately positive results of both tests require invasive diagnostic methods such as colonoscopy or bronchoscopy to confirm cancer occurrence. Nevertheless, the epigenetic based diagnostic tests have not yet been applied in routine diagnostics also due to the availability of very few results of prospective clinical trials, which could confirm the utility of such markers in a daily practice. Moreover, lack of elaborated recommendations or guidelines to carry out diagnostics with their usage limits the estimation of their presence in cancer patients. Consequently this also raises doubts regarding the technique which should be preferred for material collection and finally what material is the most valuable for methylation screening? Despite biomarker validation, the each step of a diagnostic procedure (from sample collection to molecular analysis) also needs to be validated. The potential algorithm of markers selection and validation into diagnostics is presented in Figure 1.
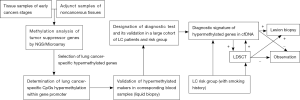
It seems that in nearly future the subsequent genes will be examined as potential lung cancer biomarker. Based on currently achieved findings the most promising is selection of tumor-specific hypermethylated genes and combination these into diagnostic panel. Such proceeding could significantly improve accuracy of designed molecular tests. Strongly recommended also seems to be combining of methylation analysis with analysis of other epigenetic alterations such as microRNA expression which potential utility in lung cancer screening was proven in COSMOS and MILD trials. All recent findings indicate that analysis of methylation status of tumor suppressor genes promoters will not be a single diagnostic tool that allows early diagnosis of lung cancer but rather will be applied as adjunct test for imaging diagnostics. Such application of epigenetic tests significantly reduce false results in LDCST examination and prevent patients form unnecessary surgery or invasive biopsy what is always disturbing and stressful. Finally, the sputum examination should not be underestimated in cancer detection and may be used as complementary examination for serum/plasma analysis when the results are inconclusive. Thanks to possibility to examination of methylation status with non-invasive manner (liquid biopsy, sputum collection), analysis of such epigenetic alteration will be one of a leading priority in lung cancer early detection.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Shaohua Cui (Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.12.51). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 2012;13:607-15. [Crossref] [PubMed]
- Maisonneuve P, Bagnardi V, Bellomi M, et al. Lung cancer risk prediction to select smokers for screening CT--a model based on the Italian COSMOS trial. Cancer Prev Res (Phila) 2011;4:1778-89. [Crossref] [PubMed]
- Orłowski T. Early lung cancer—the role of screening programs. Pneumonol Alergol Pol 2014;82:1-2. [PubMed]
- Brunelli A, Charloux A, Bolliger CT, et al. ERS/ESTS clinical guidelines on fitness for radical therapy in lung cancer patients (surgery and chemo-radiotherapy). Eur Respir J 2009;34:17-41. [Crossref] [PubMed]
- Sculier JP, Berghmans T, Meert AP. Advances in target therapy in lung cancer. Eur Respir Rev 2015;24:23-9. [Crossref] [PubMed]
- Ru Zhao Y, Xie X, de Koning HJ, et al. NELSON lung cancer screening study. Cancer Imaging 2011;11 Spec No A:S79-84.
- Saghir Z, Dirksen A, Ashraf H, et al. CT screening for lung cancer brings forward early disease. The randomised Danish Lung Cancer Screening Trial: status after five annual screening rounds with low-dose CT. Thorax 2012;67:296-301. [Crossref] [PubMed]
- Wen J, Fu J, Zhang W, et al. Genetic and epigenetic changes in lung carcinoma and their clinical implications. Mod Pathol 2011;24:932-43. [Crossref] [PubMed]
- Mehta A, Dobersch S, Romero-Olmedo AJ, et al. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev 2015;34:229-41. [Crossref] [PubMed]
- Phillips T. The role of methylation in gene expression. Nature Education 2008;1:116.
- Siegfried Z, Simon I. DNA methylation and gene expression. Wiley Interdiscip Rev Syst Biol Med 2010;2:362-71. [Crossref] [PubMed]
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011;2:607-17. [Crossref] [PubMed]
- Chédin F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci 2011;101:255-85. [Crossref] [PubMed]
- Brock G, Castellanos-Rizaldos E, Hu L, et al. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res 2015;4:280-90.
- Bedard PL, Hansen AR, Ratain MJ, et al. Tumour heterogeneity in the clinic. Nature 2013;501:355-64. [Crossref] [PubMed]
- Humphrey LL, Teutsch S, Johnson M, et al. Lung cancer screening with sputum cytologic examination, chest radiography, and computed tomography: an update for the U.S. Preventive Services Task Force. Ann Intern Med 2004;140:740-53. [Crossref] [PubMed]
- Shivapurkar N, Stastny V, Suzuki M, et al. Application of a methylation gene panel by quantitative PCR for lung cancers. Cancer Lett 2007;247:56-71. [Crossref] [PubMed]
- Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res 2006;66:3338-44. [Crossref] [PubMed]
- Belinsky SA, Klinge DM, Dekker JD, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res 2005;11:6505-11. [Crossref] [PubMed]
- Shivapurkar N, Stastny V, Xie Y, et al. Differential methylation of a short CpG-rich sequence within exon 1 of TCF21 gene: a promising cancer biomarker assay. Cancer Epidemiol Biomarkers Prev 2008;17:995-1000. [Crossref] [PubMed]
- Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 2014;1846:539-46.
- González-Masiá JA, García-Olmo D, García-Olmo DC. Circulating nucleic acids in plasma and serum (CNAPS): applications in oncology. Onco Targets Ther 2013;6:819-32. [PubMed]
- Ulivi P, Silvestrini R. Role of quantitative and qualitative characteristics of free circulating DNA in the management of patients with non-small cell lung cancer. Cell Oncol (Dordr) 2013;36:439-48. [Crossref] [PubMed]
- Begum S, Brait M, Dasgupta S, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res 2011;17:4494-503. [Crossref] [PubMed]
- Hsu HS, Chen TP, Hung CH, et al. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 2007;110:2019-26. [Crossref] [PubMed]
- Hulbert A, Jusue Torres I, Stark A, et al. Early detection of lung cancer using DNA promoter hypermethylation in plasma and sputum. Clin Cancer Res 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Wielscher M, Vierlinger K, Kegler U, et al. Diagnostic performance of plasma DNA methylation profiles in lung cancer, pulmonary fibrosis and COPD. EBioMedicine 2015;2:929-36. [Crossref] [PubMed]
- Weiss G, Schlegel A, Kottwitz D, et al. Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J Thorac Oncol 2017;12:77-84. [Crossref] [PubMed]
- Powrózek T, Krawczyk P, Kuźnar-Kamińska B, et al. Analysis of RTEL1 and PCDHGB6 promoter methylation in circulating-free DNA of lung cancer patients using liquid biopsy: A pilot study. Exp Lung Res 2016;42:307-13. [Crossref] [PubMed]
- Zhu CS, Pinsky PF, Kramer BS, et al. The prostate, lung, colorectal, and ovarian cancer screening trial and its associated research resource. J Natl Cancer Inst 2013;105:1684-93. [Crossref] [PubMed]
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Kramer BS, Berg CD, Aberle DR, et al. Lung cancer screening with low-dose helical CT: results from the National Lung Screening Trial (NLST). J Med Screen 2011;18:109-11. [Crossref] [PubMed]
- Bach PB, Mirkin JN, Oliver TK, et al. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA 2012;307:2418-29. [Crossref] [PubMed]
- Hubers AJ, Heideman DA, Duin S, et al. DNA hypermethylation analysis in sputum of asymptomatic subjects at risk for lung cancer participating in the NELSON trial: argument for maximum screening interval of 2 years. J Clin Pathol 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Gu C, Lu J, Cui T, et al. Association between MGMT promoter methylation and non-small cell lung cancer: a meta-analysis. PLoS One 2013;8:e72633 [Crossref] [PubMed]
- Deep JS, Sidhu S, Chandel A, et al. Aberrant Methylation in Promoters of GSTP1, p16, p14, and RASSF1A Genes in Smokers of North India. ISRN Pulmonology 2012;e247631
- Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. [Crossref] [PubMed]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. RARβ2 gene methylation level in the circulating DNA from blood of patients with lung cancer. Eur J Cancer Prev 2011;20:453-5. [Crossref] [PubMed]
- Powrózek T, Krawczyk P, Nicoś M, et al. Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin Transl Oncol 2016;18:398-404. [Crossref] [PubMed]
- Nie K, Jia Y, Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol 2015;36:7-19. [Crossref] [PubMed]
- Powrózek T, Krawczyk P, Kucharczyk T, et al. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Med Oncol 2014;31:917. [Crossref] [PubMed]
- Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci 2015;2:13. [Crossref] [PubMed]


