
Molecular characterization of short-term primary cultures and comparison with corresponding tumor tissue of Brazilian glioblastoma patients
Introduction
Central nervous system (CNS) tumors account for approximately 2% of all adult malignancies worldwide and represent the most prevalent cancer among those 0–19 years old (1,2). The SEER (Surveillance, Epidemiology, and End Results Program) cancer database recorded an incidence of 6.4 per 10,000 persons per year and 4.3 per 10,000 persons per year during 2008–2012 in the United States (3). In Brazil, there are expected to be 5,540 new cases of CNS tumors in men and 4,830 in women in 2016, making them the eleventh most frequent tumor type, considering both genders (4). Among the malignant CNS tumors, gliomas are the most frequent and are divided into four WHO malignancy grades (5). Glioblastoma (WHO grade IV) is not only the most aggressive but is also the most frequent subtype, representing more than 50% of all adult gliomas (2,5-7). Clinically, these tumors can be subdivided into primary or de novo glioblastoma, which appears in older patients (~65 y) and without clinical or histopathological evidence of a preexisting lesion, or secondary glioblastoma, arising in young adult patients (~45 y) from lower grade lesions, such as diffuse astrocytoma (WHO grade II) or anaplastic astrocytoma (WHO grade III) (5,8-10). Histologically, glioblastomas are characterized by high cellularity, diffuse infiltration, intense mitotic activity, microvascular proliferation, nuclear atypia, cellular pleomorphism, necrosis and a high intertumoral and intratumoral morphological heterogeneity, which demonstrate the high genomic instability of these tumors (5,10,11). Glioblastoma is one of the most devastating human diseases, with a mean survival of 12 to 15 months (5,12-14). Currently, the standard treatment consists in the widest possible resection followed by a combined regimen of radiation therapy and adjuvant temozolomide-based chemotherapy (15,16).
In the last decade, recurrent genomic aberrations of glioblastomas have been elucidated, and it is now clear that several molecular pathways distinguish adult from pediatric cases and primary from secondary glioblastomas, and recently, 5 molecular subclasses (proneural G-CIMP, proneural non G-CIMP, neural, classical and mesenchymal) were reported in adult glioblastomas (10,13,17-19). According to The Cancer Genome Atlas (TCGA), the tumor suppressor genes PTEN and TP53 are among the most frequently mutated genes, especially among newly diagnosed adult patients (14,17). The integrated analysis of the complex chromosomal and genetic aberrations present in glioblastomas shows an interaction between three major signaling pathways: the receptor tyrosine kinase receptors (RTKs), the p53 and RB (14,17). In fact, 74% of glioblastomas harbor somatic alterations of these pathways, suggesting that their deregulation is extremely important for tumor development (14,17). An analysis by the TCGA consortium confirmed that losses are more frequent than amplifications in GBM (17,20). Brennan et al. analyzed the DNA copy-number alterations of 543 glioblastomas and showed that the amplifications and copy number gains are more frequent on chromosomes 7 (7p11.2, 7q31.2 and 7q21.2), 12 (12q14.1 and 12q15) and 4 (4q12), and the main deletions occur on chromosomes 10 (10q23.31), 13 (13q14.2 and 13q21.2), 22 (22q13.31) and 9 (9p21.3) (17,21). In addition, mutations in the IDH1/2, PDGFRA and NF1 genes have been associated with the development of glioblastoma (22). Notably, several recent studies report a high frequency (50–83%) of mutations in the promoter region of the TERT gene, constituting one of the most common somatic events in adult glioblastomas currently observed (23-25).
Despite these major advances in the understanding of the genomics of glioblastoma, there has been little progress in developing therapeutic applications, so the clinical impact has been limited. For this reason, it is essential to develop model systems that better mimic the genomic diversity of individual tumors and use these systems for preclinical research designed to develop better treatment options (26). One of these models is the development of short-term primary cell cultures from patient tumors, which has become more popular in recent years and has the practical advantage of providing an unrestricted source of material for tumor biology and for the characterization and testing of new drugs (26,27).
In the present study, we aimed to determine the molecular profile of short-term glioblastoma primary cultures using array-CGH (aCGH), next-generation sequencing (Ion Torrent) and Sanger sequencing for confirmations and extended the analysis to the TERT promoter gene. We also compared the molecular profiles of the short-term primary cultures with the corresponding frozen tumor tissues.
Methods
Samples
Four short-term tumor primary cultures were established from patients diagnosed with glioblastoma (WHO grade IV) at Barretos Cancer Hospital, Barretos, São Paulo, Brazil. The clinico-pathological characteristics, such as tumor localization, resection, treatment, KPS (Karnofsky Performance Status) and patient status, were collected from medical records. The resected tumor was divided into three portions. The first portion was sent to the Pathology department for a confirmatory diagnosis, the second piece was frozen and stored at a hospital biobank, and the last portion was used for primary cell culture establishment. All of the patients recruited in the study gave informed consent, and the experimental protocols were approved by the Barretos Cancer Hospital local Ethics Committee (491/2011).
Establishment of short-term primary cell cultures
The primary cultures were established from resected glioblastomas, which were pre-operatively detected by magnetic resonance imaging (MRI). In the operating room, the excised tumor tissue was immediately placed in 1X PBS (Phosphate Buffer Solution-Sigma-Aldrich) and was sent to the laboratory to be processed as previous described by Martinho et al. (28), within a period that ranged from 30 minutes (RT, room temperature) to a maximum of 24 hours (4 °C). Briefly, the fresh tumor tissues were washed in 1X PBS to remove the remaining blood and debris of the tumor tissue. After washing, the tissue was minced with forceps and a scalpel, and the blood vessels and apparent necrotic regions were removed. A trypsin solution (0.01%) (Gibco, Invitrogen) was added to the small pieces of tissue, followed by incubation at 37 °C for 30 minutes for the total digestion of the tissue to obtain a viable cell suspension. During this incubation, the minced sample was gently pipetted to aid cellular disaggregation. The viable cell suspension was then placed in a T25 flask containing DMEM (Dulbecco’s Modified Eagle Medium-Sigma-Aldrich) supplemented with 10% FBS (Fetal Bovine Serum-Sigma-Aldrich) and 1% PS (Penicillin-Streptomycin-Invitrogen) at 37 °C in a humidified atmosphere of 5% carbon dioxide in air. The medium was renewed every 48 or 72 hours, depending on the cell density and the growth kinetics. The primary cultures were considered established short-term culture when they reached ten passages.
Immunocytochemistry
Approximately 1.0×106 cells were seeded on 15 mm circular coverslips and were incubated in DMEM supplemented with 10% FBS and 1% PS at 37 °C in a humidified atmosphere of 5% carbon dioxide in air until they reached confluence. After that, the cells were fixed with 4% paraformaldehyde (Merck) and were permeabilized with 100% Triton-X (Merck). Immunocytochemistry was performed using the streptavidin-biotin-peroxidase complex system (UltraVision Large Volume Detection System Anti-Polyvalent, HRP; LabVision Corporation) according manufacturer’s instructions. Anti-GFAP (clone Z00334, DAKO) was used at a concentration of 1:1,000, and Anti-Nestin (clone 10C2, Novus Biologicals) was used at a concentration of 1:200. The chromogen DAB (3, 3’-Diaminobenzidine) was used to detect the immune reaction. The images were acquired by an Olympus Optical Microscope (magnification 100×).
DNA isolation
For further molecular analysis, DNA was isolated from 1.0×106 tumor cells in the third passage (3P) using TRIzol® (Life Technologies) according to the manufacturer’s instructions. For the frozen tumors, the tissue was macrodissected, ensuring more than 75% tumor cell content, and DNA (from tumor and blood) was isolated using a DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instructions. All of the DNA samples were quantified by a NanoDrop® 2000 (Thermo Scientific) and a Qubit® 2.0 Fluorometer (Life Technologies) and were then stored at −20 °C for genetic analysis.
Identification of the short tandem repeat (STR) profiles of the primary cultures
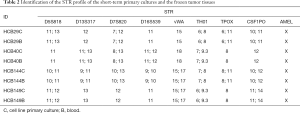
The STR profiles were performed in the established short-term primary cultures and were further compared with the corresponding patient blood DNA using a panel of eight STR loci (D5S818, D13S317, D7S820, D16S539, vWA, TH01, TPOX and CSF1P0) plus gender determination (AMEL), as previously reported (29,30) .
aCGH and data analysis
All four of the short-term primary cultures and respective frozen tumors were assessed by aCGH to evaluate the chromosomal alterations using the Agilent Human Genome CGH 8×60 K arrays following the manufacturer’s instructions (Agilent Technologies-version 7.1) as previously described by our group (31). The copy number alterations (CNA) were evaluated by a comparison with the copy number found in the reference DNA (DNA universal control-Promega Madison WI USA-Male Reference: G147A; Woman Reference: G152A).
The txt files generated by the Feature Extraction software, containing the intensity measurements for all of the spots were imported into a script written in R (32) for the bioinformatics analysis as previously described (31). The data were deposited under GEO Number GSE59967.
Ion Torrent sequencing and data analysis
The four short-term primary cell cultures and the frozen tumor tissues were sequenced using the Ion AmpliSeq™ Cancer Panel Primer Pool (Life Technologies), which analyzed the presence of mutations in 46 cancer related genes. The protocol was carried out according to the manufacturer’s instructions (Life TechnologiesTM) and as previously reported by our group (33).
All of the analyses from the Ion Torrent sequencing were performed on the Torrent Server (Life TechnologiesTM).
Sanger sequencing
The analysis of the hotspot mutations of TP53 (exons 5–8), BRAF (exon 15), IDH1 (exon 4), IDH2 (exon 4), FGFR1 (exons 12 and 14), the TERT promoter region (hotspots -124 bp and -146 bp) and the PTEN gene (exons 1–9) was performed by PCR followed by direct sequencing. The primers and PCR conditions of TP53, BRAF, IDH1, IDH2, FGFR1, TERT and PTEN were previously described (34-39).
The PCR products were then submitted to direct sequencing using an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit and were analyzed on a Genetic Analyzer ABI PRISM® 3500 (Applied Biosystems). The electropherograms were compared to GenBank (TP53: ENSG00000141510; PTEN: ENSG00000171862; BRAF: ENSG00000157764; IDH1: ENSG00000138413; IDH2: ENSG00000182054; H3F3A: ENSG00000163041; FGFR1: ENSG00000077782; TERT: ENSG00000164362). All of the samples that presented any alterations were confirmed with at least two additional independent amplifications and sequencing reactions.
Integration of chromosomal and mutation profiles
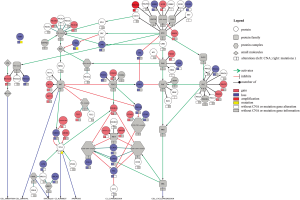
To perform the integration of chromosomal and mutation data and to assess the signaling pathway alterations, we input the CNA and mutations found in the Mondrian software plugin (http://cbio.mskcc.org/mondrian/) in the Cytoscape network visualization software (http://cytoscape.org) using the data from one of the primary cultures as an example.
Statistical analysis
The statistical analysis was performed using SPSS software (Statistical Package for Social Science version 19.0). The characterization of the Brazilian population in this study, including the clinico-pathological and molecular data, was performed using descriptive statistics (mean and frequencies).
Results
Establishment, success rates and immunocytochemistry characterization of short-term glioblastoma primary cultures
From February 2011 to December 2012, 67 primary glioma tumors were collected to obtain short-term primary cell cultures, and from these, 53.73% (36/67) successfully attached and grew. The cell lines were considered established short-term cultures when growing cultures attained at least 10 passages without necessarily having been frozen and thawed. From the 36 established short-term glioblastoma primary cell cultures, the four with the most rapid and stable growth kinetics were characterized in more detail.
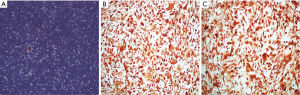
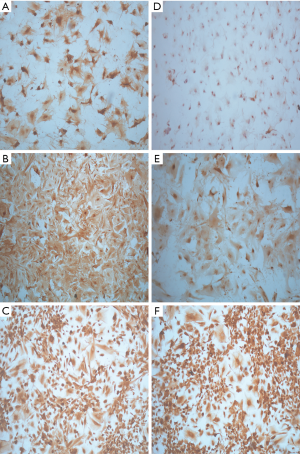
All four of the short-term glioblastoma primary cultures exhibited an attached, monolayer culture growth and showed a homogeneous morphology with a fibroblast-like phenotype characteristic of glial cells (Figure 1A). The growth pattern or time until confluence of the short-term primary cell cultures that reached the first passage (1P) varied between 7 to 10 days. Following this period, the attached and growing cells were passaged two more times and were characterized for GFAP expression. All of the cell lines showed strong GFAP expression (Figure 1B and Figure S1A-C), and in accordance with the formalin-fixed paraffin-embedded primary tumors, GFAP immunostaining was analyzed in the Pathology department of Barretos Cancer Hospital for diagnostic purposes (data not shown). Similarly, all of the cell lines showed a predominantly strong Nestin expression (Figure 1C and Figure S1D-F).
Clinico-pathological features of glioblastomas
The clinico-pathological features tumor localization, KPS, resection, patient status, and radio- and chemotherapy of the patients are summarized in Table 1. The mean age of the patients enrolled in this study was 57.75 years (ranging from 53 to 64) and the overall survival (OS) rate was 5.75±3.4 months (mean ± SEM) (Table 1). The STR profile of the four short-term primary cell lines and the corresponding blood DNA confirmed that the short-term primary cell lines were derived from the corresponding patients (Table 2).

Full table
Full table
aCGH profile
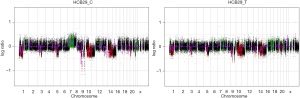
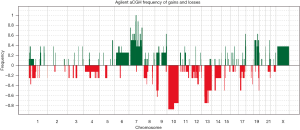
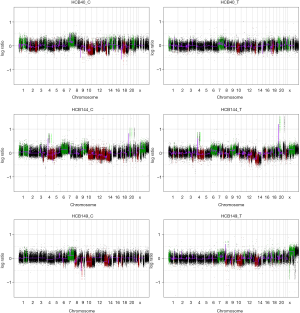
aCGH was performed in the four short-term primary cell cultures and the four corresponding frozen tumor tissues to evaluate the chromosomal abnormalities (Figure 2 and Figure S2). A summary of all of the DNA CNA detected in both the primary culture and the frozen tumor tissue is shown in Table 3. An overview of the CNA identified in the primary cell lines is represented in Figure 3. Amplifications were observed in the HCB144 cell line, and deletions were identified on the HCB29, HCB40 and HCB149 cell lines (Figure 2, Figure S2 and Table 3). In the HCB144 cell line, the amplified 4q11-q12 region harbored the PDGFRA, KIT and VEGFR2 oncogenes. The aCGH analysis showed that the most frequent events affected chromosomes 7, 10, 13, 17 and 19 (Figure 3). Gain of chromosome 7 was present in up to 80% of the cases, with a minimum gain region restricted to 7p12.1–q11.23, followed by gain of chromosome 19 (approximately 50%). Loss of chromosome 10 was found in all of the cases, and the region 10p15.3–q26.3 was altered in all of the primary cell lines, followed by loss of chromosome 13 (approximately 60%) and loss of 17p (occurred in 40%) (Figure 3). The major CNAs were chromosomal losses with 82 events, followed by gains (79 events), and, less frequently, amplifications and deletions (Table 3).

Full table
Furthermore, the aCGH revealed that the 9p22.1–p21.3 region was deleted in two of the cell lines (HCB29 and HCB149) and involved the MTAP, C9orf53 and CDKN2A genes (Table 3). In HCB40, we observed a deletion of the 17q11 region, harboring the POLDIP2 gene (Table 3).
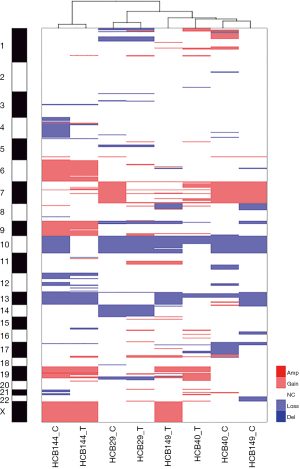
When we compared the CNA profiles of the short-term primary cell lines with the chromosomal alterations identified in the corresponding frozen glioblastoma tissues, the genomic profiles were very similar (Figure 2 and Table 3). These findings were further corroborated by an unsupervised hierarchical cluster analysis, which showed close clustering of the paired primary culture and frozen tissue, with the exception of the HCB149 samples (Figure 4). Nevertheless, we also observed that some chromosomal aberrations were exclusively found in either the primary cell culture or the frozen tumor tissue (Table 3, Figure 2 and Figure 4).
Mutation profile
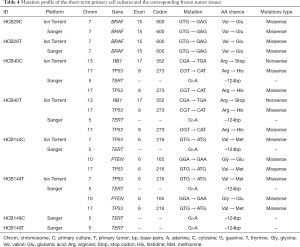
To determine the overall somatic mutational profiles in the glioblastoma study group, all of the short-term primary cell cultures and the frozen tumor DNA and blood DNA were sequenced initially performed using the Ion Torrent platform. We observed oncogenic mutations in the BRAF, RB1 and TP53 genes, as shown in Table 4. To validate the mutation status identified by Ion Torrent in a subset of genes (TP53, BRAF, FGFR1, IDH1, IDH2 and PTEN) and to extend to other important genes (TERT promoter region) not included in the AmpliSeq panel, we performed Sanger sequencing of the hotspot regions of those genes (Table 4). With the exception of the PTEN mutation (p.Gly165Glu), both Sanger and Ion Torrent identified the same alterations. This PTEN mutation was in a codon that was not covered by the AmpliSeq library. The most frequently mutated gene was the TERT promoter, which was found in 3 of the 4 short-term primary cell lines, followed by TP53 mutations in 2 of the 4 short-term primary cultures. Additionally, we found mutations in the BRAF and RB1 genes. No mutations were detected in the IDH1, IDH2, and FGFR1 genes.
Full table
As shown in Table 4, all of the mutations observed in the short-term primary cell lines, except for PTEN, as shown above, were also found in the frozen tumor tissues, confirming that the cell culture environment did not lead to acquisition of additional somatic mutations.
Discussion
To gain a better understanding of the biology of glioblastoma and to contribute to future functional and therapeutic studies, we obtained short-term glioblastoma primary cultures and characterized their chromosomal and molecular alterations. We showed that the four short-term glioblastoma cell cultures harbored a genomic profile that was similar to the corresponding matched frozen tumors, suggesting that they might constitute a reliable model for more detailed preclinical studies.
The success rate in the establishment of the short-term primary cell tumors varies greatly among studies (40-42). Several methodologies have been used for the establishment of primary glioma cell lines (26,40). Some authors use growth factors, such as bFGF and EGF, without serum, whereas others used serum-cultured conditions (41,42). The presence of serum in the culture is described to induce the differentiation and expansion of neoplastic cells, potentially eliminating the glioma stem cells (41-43). Lee et al. reported that glioblastoma primary cell lines grown without serum have the capability to form neurospheres in vitro, have a vast potential to self-renew, undergo multilineage differentiation, have genetic stability over serial passages and harbor all of the alterations found within primary tumors, while the same cells cultured under serum conditions lose these abilities and change their gene expression profiles (42). Similarly, Behnan et al. studied glioblastoma primary cultures under different conditions, such as neurospheres and adherent cell lines without and with serum (43). They observed that glioblastoma cell lines in serum conditions lose their tumorigenicity potential after the removal the non-adherent cells, but when grown in a mixed condition, they express both phenotypes, tumorigenic and non-tumorigenic profiles, suggesting that the serum does not preclude the glioblastoma cells’ tumorigenicity since the cell culture condition allows the growth of the tumorigenic cells (43). We and others used serum-containing medium, and the subsequent molecular analysis showed that short-term primary cultures exhibited a profile that was similar to their tumor tissue counterparts (27,28,40). Therefore, despite this putative limitation in tumor cell culture that is not enriched for the heterogeneity of tumor cell type of glioblastomas, we observed identical genetic alterations.
The copy number analysis of all four of the glioblastoma cell lines showed that the most prevalent alterations affected chromosomes 7, 10, 13, 17 and 19, confirming previous studies in glioblastoma, including those from our group (13,31,44,45). Brennan et al. (17) and Beroukhim et al. (20) observed that chromosomal losses were more frequent than amplifications, as shown in this study. The amplification of the important region 4q12, which harbors the KIT, PDGFRA and KDR oncogenes, was observed in one case. This finding is in agreement with other studies that showed a portion of adult glioblastomas with co-amplification of these genes (17,22,28,46-48). It is important to note that PDGFRA amplification is more common in pediatric glioblastoma or diffuse intrinsic pontine gliomas (DIPG) than in adult glioblastoma (22,31). The deletion of the region 9p22.1–21.3, involving the CDKN2A/B, MTAP and C9orf53 genes, was seen in half of the established short-term cell lines, which is in accordance with previous studies (12,17,44,49). Importantly, the tumor suppressor gene CDKN2 is involved in the development of glioblastoma, with a high-level homozygous deletion (12,17). This finding also corroborates the integrated analysis of TCGA that showed that the RB pathway is one of the most altered in gliomagenesis (14,17). TCGA_2008 (14) showed that an integrated analysis of the complex chromosomal and genetic aberrations in glioblastoma provides an overall evaluation of the biology of glioblastoma and points toward potential therapeutic strategies. Using a similar approach, we observed that most of the alterations occur in these three signaling pathways, including the RTKs and p53 and RB, in concordance with TCGA data (Figure S3). Although the importance of MTAP in gliomas has not been well studied, this deletion has been reported with high frequency in glioblastoma (39,50,51). Another deleted region was 17q11.2, which is composed of the POLDIP 2 gene, or POLD2. The role of the POLDIP 2 gene in glioblastoma is very poorly characterized, with just a study suggesting the association with survival patients (52). However, we could not dispute its importance, since it is related to DNA replication and repair, besides being regulated by PTEN (52). Furthermore, the comparison of the CNA profiles of the short-term primary cell lines with respective frozen tumor tissues showed that they clustered together, with concordant chromosomal copy number profiles. The unique short-term culture and the frozen tumor tissue that did not cluster together was HCB149. Nevertheless, minor alterations found uniquely either in short-term primary cell culture or in frozen tumor tissue were detected. Such small discrepancies between the frozen and cultured tumors can be justified by the enrichment of the tumor cells in the cell culture environment and have also been reported by other studies (21,40).
The evaluation of the mutation profile using the AmpliSeq panel (IonTorrent platform) showed the presence of oncogenic mutations on the BRAF, RB1 and TP53 genes. The RB1 gene mutations are reported to be highly frequent in primary glioblastoma (13); however, we could not observe this high frequency in the analyzed samples, which was probably due to our small sample size. The findings of the TP53 (2/4) and BRAF (1/4) genes, although not the most expected in primary glioblastomas, are in concordance with some studies also reporting the presence of these alterations in primary glioblastoma (14,53). However, it is important to highlight that TP53 and BRAF gene mutations are more frequent in secondary (12,54) and pediatric glioblastomas (18). To validate these results, we performed Sanger sequencing of select mutated genes and evaluated the additional mutation status of the hotspot in the TERT promoter region, which has been reported as a major player in the development of glioblastoma (50,55). As expected, we found a high frequency (75%) of this hotspot TERT promoter mutation (c.-124C > T), in concordance with recent studies (23,24,56). TERT promoter mutations have been extensively associated with malignant development, including glioblastoma, due to its capacity to induce telomerase reactivation (57,58).
As expected, no mutations were seen in the IDH1 and IDH2 genes, which is common in secondary glioblastomas (59), and none were seen in FGFR1, which is frequent in pediatric tumors (25). All of the mutations were identified in both the short-term primary cell cultures and the frozen tumor tissues, indicating that the alterations were not induced by the cell culture environment.
Conclusions
In conclusion, the present findings showed the successful establishment of short-term glioblastoma primary cultures, which exhibited chromosomal and genetic alterations in regions previously associated with glioblastoma tumorigenesis. Moreover, these glioblastoma cultures demonstrated good reproducibility and concordance between the genomic profiles of the short-term primary cell cultures and the respective frozen tumor tissues, indicating their utility for preclinical studies of glioblastoma biology.
Acknowledgments
The authors acknowledge Flávia Escremim and Adriane Feijó Evangelista for technical collaboration in this study.
Funding: This study was supported by the Universal/CNPq (475358/2011-2-Reis RM), FAPESP (2012/19590-0-Reis RM) and the MCTI/CNPq No. 73/2013 (Reis RM) grants. Bidinotto LT was a recipient of the FAPESP fellowship (2011/08523-7 and 2012/08287-4).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.32). RMR serves as an unpaid editorial board member of Translational Cancer Research from Nov 2016 to Oct 2018. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The experimental protocols were approved by the Barretos Cancer Hospital local Ethics Committee (491/2011) and all of the patients recruited in the study gave informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cancer Incidence and Mortality Worldwide. International Agency for Research on Cancer: IARC Cancer Database, 2016. Available online: http://globocan.iarc.fr/Pages/fact_sheets_population.aspx
- Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol 2014;16:iv1-63. [Crossref] [PubMed]
- SEER Cancer Statistics Review, 1975-2012. National Cancer Institute, 2016. Available online: http://seer.cancer.gov/csr/1975_2012
- Estimativa 2016 Incidência de Câncer no Brasil. Instituto Nacional de Câncer José de Alencar Gomes da Silva-INCA. 2015, 2016. Available online: http://www.inca.gov.br/wcm/dncc/2015/por-tipos.asp
- Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97-109. [Crossref] [PubMed]
- Dolecek TA, Propp JM, Stroup NE, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol 2012;14:v1-49. [Crossref] [PubMed]
- Vivanco I, Robins HI, Rohle D, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov 2012;2:458-71. [Crossref] [PubMed]
- Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007;21:2683-710. [Crossref] [PubMed]
- Scherer HJ. A critical review: the pathology of cerebral gliomas. J Neurol Psychiatry 1940;3:147-77. [Crossref] [PubMed]
- Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res 2013;19:764-72. [Crossref] [PubMed]
- Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A 2012;109:3041-6. [Crossref] [PubMed]
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73. [Crossref] [PubMed]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008;359:492-507. [Crossref] [PubMed]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [Crossref] [PubMed]
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987-96. [Crossref] [PubMed]
- Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 2011;29:4482-90. [Crossref] [PubMed]
- Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462-77. [Crossref] [PubMed]
- Sturm D, Bender S, Jones DT, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer 2014;14:92-107. [Crossref] [PubMed]
- Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006;9:157-73. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Xie Y, Bergström T, Jiang Y, et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine 2015;2:1351-63. [Crossref] [PubMed]
- Aldape K, Zadeh G, Mansouri S, et al. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol 2015;129:829-48. [Crossref] [PubMed]
- Nonoguchi N, Ohta T, Oh JE, et al. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol 2013;126:931-7. [Crossref] [PubMed]
- Vinagre J, Almeida A, Pópulo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun 2013;4:2185. [Crossref] [PubMed]
- Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 2013;110:6021-6. [Crossref] [PubMed]
- Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer 2010;10:241-53. [Crossref] [PubMed]
- Huveldt D, Lewis-Tuffin LJ, Carlson BL, et al. Targeting Src family kinases inhibits bevacizumab-induced glioma cell invasion. PLoS One 2013;8:e56505 [Crossref] [PubMed]
- Martinho O, Silva-Oliveira R, Miranda-Gonçalves V, et al. In Vitro and In Vivo Analysis of RTK Inhibitor Efficacy and Identification of Its Novel Targets in Glioblastomas. Transl Oncol 2013;6:187-96. [Crossref] [PubMed]
- Dirks WG, Drexler HG. Online verification of human cell line identity by STR DNA typing. Methods Mol Biol 2011;731:45-55. [Crossref] [PubMed]
- Silva-Oliveira RJ, Silva VA, Martinho O, et al. Cytotoxicity of allitinib, an irreversible anti-EGFR agent, in a large panel of human cancer-derived cell lines: KRAS mutation status as a predictive biomarker. Cell Oncol (Dordr) 2016;39:253-63. [Crossref] [PubMed]
- Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol 2010;28:3061-8. [Crossref] [PubMed]
- Team RC. R: A language and environment for statistical computing Vienna 2012. Available online: http://www.R-project.org/
- Bidinotto LT, Scapulatempo-Neto C, Mackay A, et al. Molecular Profiling of a Rare Rosette-Forming Glioneuronal Tumor Arising in the Spinal Cord. PLoS One 2015;10:e0137690 [Crossref] [PubMed]
- Reis RM, Könü-Lebleblicioglu D, Lopes JM, et al. Genetic profile of gliosarcomas. Am J Pathol 2000;156:425-32. [Crossref] [PubMed]
- Basto D, Trovisco V, Lopes JM, et al. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathol 2005;109:207-10. [Crossref] [PubMed]
- Tefferi A, Lasho TL, Abdel-Wahab O, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 2010;24:1302-9. [Crossref] [PubMed]
- Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22:425-37. [Crossref] [PubMed]
- Liu X, Wu G, Shan Y, et al. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle 2013;12:1637-8. [Crossref] [PubMed]
- Sasaki T, Arai H, Beppu T, et al. Detection of gene amplification and deletion in high-grade gliomas using a genome DNA microarray (GenoSensor Array 300). Brain Tumor Pathol 2003;20:59-63. [Crossref] [PubMed]
- Mullins CS, Schneider B, Stockhammer F, et al. Establishment and characterization of primary glioblastoma cell lines from fresh and frozen material: a detailed comparison. PLoS One 2013;8:e71070 [Crossref] [PubMed]
- Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009;4:568-80. [Crossref] [PubMed]
- Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006;9:391-403. [Crossref] [PubMed]
- Behnan J, Stangeland B, Hosainey SA, et al. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene 2017;36:570-84. [Crossref] [PubMed]
- Baronchelli S, Bentivegna A, Redaelli S, et al. Delineating the cytogenomic and epigenomic landscapes of glioma stem cell lines. PLoS One 2013;8:e57462 [Crossref] [PubMed]
- Sintupisut N, Liu PL, Yeang CH. An integrative characterization of recurrent molecular aberrations in glioblastoma genomes. Nucleic Acids Res 2013;41:8803-21. [Crossref] [PubMed]
- Martinho O, Longatto-Filho A, Lambros MB, et al. Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas. Br J Cancer 2009;101:973-82. [Crossref] [PubMed]
- Paugh BS, Zhu X, Qu C, et al. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res 2013;73:6219-29. [Crossref] [PubMed]
- Gomes AL, Reis-Filho JS, Lopes JM, et al. Molecular alterations of KIT oncogene in gliomas. Cell Oncol 2007;29:399-408. [PubMed]
- Schwartzbaum JA, Fisher JL, Aldape KD, et al. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2006;2:494-503; quiz 1 p following 516.
- Suzuki T, Maruno M, Wada K, et al. Genetic analysis of human glioblastomas using a genomic microarray system. Brain Tumor Pathol 2004;21:27-34. [Crossref] [PubMed]
- Nakahara Y, Shiraishi T, Okamoto H, et al. Detrended fluctuation analysis of genome-wide copy number profiles of glioblastomas using array-based comparative genomic hybridization. Neuro Oncol 2004;6:281-9. [Crossref] [PubMed]
- Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA 2009;302:261-75. [Crossref] [PubMed]
- Kleinschmidt-DeMasters BK, Aisner DL, Birks DK, et al. Epithelioid GBMs show a high percentage of BRAF V600E mutation. Am J Surg Pathol 2013;37:685-98. [Crossref] [PubMed]
- Frankel RH, Bayona W, Koslow M, et al. p53 mutations in human malignant gliomas: comparison of loss of heterozygosity with mutation frequency. Cancer Res 1992;52:1427-33. [PubMed]
- Wada K, Maruno M, Suzuki T, et al. Chromosomal and genetic aberrations differ with meningioma subtype. Brain Tumor Pathol 2004;21:127-33. [Crossref] [PubMed]
- Batista R, Cruvinel-Carloni A, Vinagre J, et al. The prognostic impact of TERT promoter mutations in glioblastomas is modified by the rs2853669 single nucleotide polymorphism. Int J Cancer 2016;139:414-23. [Crossref] [PubMed]
- Kyo S, Takakura M, Fujiwara T, et al. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci 2008;99:1528-38. [Crossref] [PubMed]
- Heidenreich B, Rachakonda PS, Hemminki K, et al. TERT promoter mutations in cancer development. Curr Opin Genet Dev 2014;24:30-7. [Crossref] [PubMed]
- Smolková K, Ježek P. The Role of Mitochondrial NADPH-Dependent Isocitrate Dehydrogenase in Cancer Cells. Int J Cell Biol 2012;2012:273947 [Crossref] [PubMed]


