Association between APC promoter methylation and clinicopathological features of patients with hepatocellular carcinoma: a meta-analysis with PRISMA guideline
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor and the second most common cause of cancer-related deaths among the world (1). On the basis of global cancer statistics, an estimated 782,500 new cases were diagnosed as HCC in 2012, leading to approximately 745,500 deaths of this disease (1). Some risk factors have been showed to be involved in the development of HCC, including hepatitis B virus (HBV), hepatitis C virus (HCV) infection, liver cirrhosis, aflatoxin exposure, and alcohol consumption etc. (2-5).
Studies suggest that epigenetic alterations (DNA methylation, histone modifications, nucleosome positioning and non-coding RNAs) are an early biotic event in human cancers (6,7). As a common epigenetic change, DNA methylation plays an important role in the tumorigenesis, progression and prognosis of cancer (8-10). Mapped to human chromosome 5q21-5q22, the adenomatous polyposis coli (APC) gene, a key tumor suppressor gene (TSG), encodes a large multidomain protein and involves in the Wnt signaling pathway (11,12). Moreover, the APC gene has many biological functions, such as the regulation of cell cycle, cell migration and adhesion, transcriptional activation, apoptosis, and chromosomal instability (13-15). APC promoter methylation has been found in many human cancers, including prostate cancer (16), colorectal cancer (17), breast cancer (18), and HCC (19).
The results with respect to promoter methylation of the APC in HCC vs. benign lesions remains controversial. For example, Harder et al. reported that APC promoter had a same methylation frequency in HCC and liver cirrhosis (20). APC promoter methylation frequency was significantly higher in HCC than in liver cirrhosis by Lee et al. (21). Thus, the current meta-analysis was performed to evaluate the relationship between APC promoter methylation and HCC in cancer vs. liver cirrhosis, chronic hepatitis, and dysplastic nodules. Additionally, we also assessed the correlation between APC promoter methylation and clinicopathological features of patients with HCC, including gender, tumor stage, vascular invasion, HBV, and HCV infection status.
Methods
Search strategy
Eligible publications were identified through searching the relevant databases before December 10th, 2016, including the PubMed, Embase, EBSCO and the Cochrane Library. The search strategy was conducted based on the following search terms and key words: (APC OR adenomatous polyposis coli) AND (liver OR hepatocellular OR hepatic) AND (cancer OR tumor OR neoplasm OR carcinoma) AND (methylation OR epigene*). The reference lists from the eligible studies were carefully checked to identify other potential articles.
Inclusion criteria
Eligible articles must fulfil the following selection criteria in this meta-analysis: (I) patients were diagnosed with HCC by histopathological confirmation; (II) studies provided sufficient information to estimate the relationship between APC promoter methylation and HCC in the comparison of cancer and control groups; (III) studies provide sufficient dada to assess the correlation of APC promoter methylation with clinicopathological characteristics in HCC. When authors published several articles using the same sample population, only the most complete publication with more information were selected in our meta-analysis.
Data extraction and quality assessment
Two authors independently extracted the relevant information from the included studies: first author’s surname, year of publication, country, ethnicity, detection method of methylation, sample type, number of patients in case and control groups, methylation level, and clinicopathological parameters such as gender, tumor stage, vascular invasion, HBV, and HCV infection status. Control tissue samples included chronic hepatitis, liver cirrhosis, dysplastic nodules, and normal liver tissues. Additionally, the quality evaluation of the eligible articles was performed using the Newcastle–Ottawa Scale (NOS), ranging from 0 to 9. An individual study with a NOS score of ≥6 was considered as high quality, and a NOS score of ≤3 was considered as low quality (22,23) (Table S1).
Data analysis
The data were analyzed using Stata software (version 12.0, Stata Corporation, College Station, TX, USA). The pooled odds ratios (ORs) with their corresponding 95% confidence intervals (95% CIs) were calculated to determine whether APC promoter methylation was correlated with the risk of HCC in cancer vs. control groups. In addition, the relationship between APC promoter methylation and clinicopathological features of patients with HCC was also analyzed by the combined ORs with the corresponding 95% CIs. The chi-square test and Q statistics were applied to assess the possible heterogeneity among studies (24). The random-effects model was used in the current meta-analysis. If there was obvious evidence of heterogeneity (I2 >50% or P<0.1), a sensitivity analysis was carried out to determine the influence of the pooled OR and the change of heterogeneity by omitting an individual study (25,26). The potential publication bias was carried out using Egger’s test in cancer vs. normal liver tissue samples (27).
Results
Study characteristics
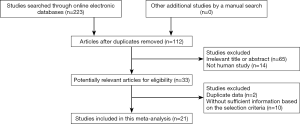
A total of 223 relevant publications were searched from a range of online electronic databases as above described (Figure 1). Based on the above selection criteria, 21 available studies (19-21,28-39,40-45) were identified in the current meta-analysis. 16 studies evaluated the relationship between APC promoter methylation and HCC in tissue samples of HCC patients vs. normal live tissues (19-21,28,30,32-35,37,38,40,41,43-45). Three studies evaluated the correlation between APC promoter methylation and HCC in blood samples of HCC patients vs. healthy blood samples (29-31). Six publications analyzed the relationship between APC promoter methylation and HCC in cancer vs. liver cirrhosis (20,21,33,37,39,44). Three publications analyzed the association between APC promoter methylation and HCC in cancer vs. dysplastic nodules (21,33,43), three publications estimated the association between APC promoter methylation and HCC in cancer vs. chronic hepatitis (21,28,37). Nine studies analyzed the correlation between APC promoter methylation and clinicopathological features of patients with HCC (31,33,35-37,39,40,42,44). All eligible articles met a score of more than or equal to 5. The basic characteristics of the included studies are presented in Table S1.
Association between APC promoter methylation and HCC in cancer vs. normal controls
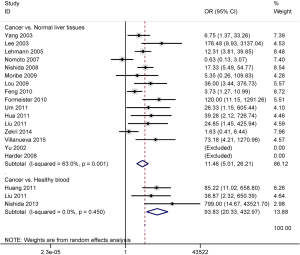
The data included the comparison of 1004 HCC patients and 212 normal live tissues, and 203 HCC patients and 109 healthy blood samples. The results demonstrated that promoter methylation of the APC gene was significantly higher in HCC than in normal live tissues and healthy blood samples (OR =11.46, 95% CI: 5.01–26.21, P<0.001; OR =93.83, 95% CI: 20.33–432.97, P<0.001; respectively) (Figure 2).
Association between APC promoter methylation and HCC in cancer vs. liver cirrhosis, chronic hepatitis, and dysplastic nodules
The result involving 296 patients with HCC and 173 patients with liver cirrhosis showed that APC promoter methylation was significantly higher in HCC than in liver cirrhosis (OR =6.04, 95% CI: 2.59–14.09, P<0.001) (Figure 3).
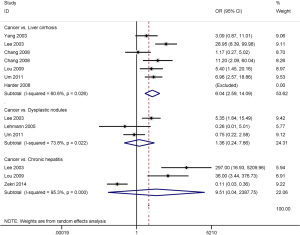
No significant correlation was observed between APC promoter methylation and HCC in cancer vs. chronic hepatitis and dysplastic nodules (OR =9.51, 95% CI: 0.04–2,387.75, P=0.424; OR =1.36, 95% CI: 0.24–7.66, P=0.728; respectively) (Figure 3), including 151 HCC patients vs. 77 patients with chronic hepatitis and 147 HCC patients vs. 74 patients with dysplastic nodules.
Subgroup analysis of APC promoter methylation in cancer vs. normal live tissues
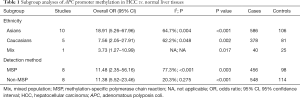
Table 1 summarizes the pooled OR based on ethnicity (Asians, Caucasians and mixed population) and detection method (MSP and non-MSP). Subgroup analysis by ethnicity showed that APC promoter methylation was associated with an increased risk of HCC in Asian, Caucasian and mixed populations (OR =18.91, 95% CI: 5.26–67.96, P<0.001; OR =7.56, 95% CI: 2.05–27.91, P=0.002; OR =3.73, 95% CI: 1.27–10.99, P=0.017; respectively).
Full table
Subgroup analysis based on detection method showed that the pooled OR of APC promoter methylation for the MSP subgroup was 11.48 (95% CI: 2.35–56.16, P=0.003), and 11.38 (95% CI: 5.52–23.46, P<0.001) for the non-MSP subgroup.
Sensitivity analysis in cancer vs. normal live tissues and liver cirrhosis
We conducted a sensitivity analysis to assess the stability of the pooled result based on the omission of one study. When cancer was compared to normal live tissues, we deleted two studies (28,41), and re-calculated the combined OR from the remaining 14 studies (OR =16.60, 95% CI: 8.36–32.96, P<0.001), with a significantly decreased heterogeneity (I2=32.5%, P=0.131).
In the comparison of cancer and liver cirrhosis, one study (21) was removed, the pooled OR from the remaining five studies was 4.40 (95% CI: 2.21–8.78, P<0.001). Meanwhile, there was no substantial evidence of heterogeneity (I2=28.1%, P=0.234).
Association between APC promoter methylation and gender, tumor stage, and vascular invasion status
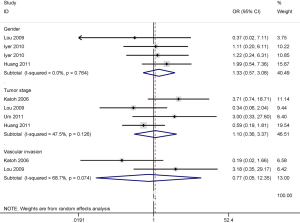
The results showed that APC promoter methylation was not correlated with gender, tumor stage, and vascular invasion status of patients with HCC (OR =1.33, 95% CI: 0.57–3.08, P=0.51; OR =1.10, 95% CI: 0.36-3.37, P=0.87; OR =0.77, 95% CI: 0.05–12.35, P=0.851; respectively), including 188, 238 and 120 HCC patients, respectively (Figure 4).
Association between APC promoter methylation and HBV and HCV infection status
The result from seven studies with 209 HCC patients demonstrated that APC promoter methylation was significantly correlated with HBV infection status of HCC patients (OR =2.86, 95% CI: 1.38–5.92, P=0.005) (Figure 5).
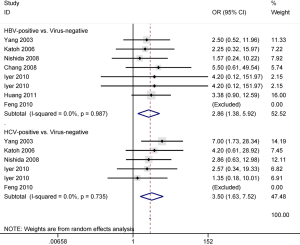
The result from five studies with 223 HCC patients showed that a significant correlation was found between APC promoter methylation and HCV infection status of HCC patients (OR =3.50, 95% CI: 1.63–7.52, P=0.001) (Figure 5).
Publication bias
Egger’s test was used to detect the potential publication bias in the comparison of cancer and normal live tissues. No obvious evidence of publication bias was found in this meta-analysis (P=0.076>0.05).
Discussion
Promoter methylation of tumor suppressor genes (TSGs) leads to the downregulation or loss of gene expression (46,47), and may markedly influence the initiation and progression of cancer (48). The reduced expression of the APC gene through promoter methylation has been found in various types of human cancers (49-51). APC promoter methylation may play a crucial role in cancer carcinogenesis and progression (52-55). The promoter of the APC gene has been shown to be frequently methylated in HCC (19,28,30). However, the clinical significance of APC promoter methylation remains unclear. We performed this meta-analysis to evaluate the association between APC promoter methylation and HCC in cancer vs. different benign lesions (liver cirrhosis, chronic hepatitis and dysplastic nodules), and the relationship of APC promoter methylation with clinicopathological characteristics of HCC patients.
Our result showed that APC promoter methylation was significantly higher in tissue samples of patients with HCC than in normal live tissues, suggesting that APC promoter methylation may play a key role in the initiation of HCC. Furthermore, subgroup analyses of ethnicity (Asian, Caucasian and mixed populations) and detection method (MSP and non-MSP) were conducted to find the difference among the different subgroups. A subgroup analysis by ethnicity revealed that APC promoter methylation was correlated with Asian, Caucasian and mixed populations with HCC, indicating that the Asian, Caucasian and mixed populations were susceptible to APC promoter methylation. Moreover, APC promoter methylation was found to be correlated with HCC risk in the MSP and non-MSP, suggesting that the MSP and non-MSP methods were sensitive to the APC gene. Interestingly, in the analysis of blood samples, the result from three studies showed that APC promoter methylation was notably higher in blood samples of HCC patients than in healthy subjects (OR =93.83, P<0.001), which indicated that APC promoter methylation may be a useful noninvasive biomarker based on blood detection. However, the results of subgroup analyses and blood samples should be carefully considered because of small sample sizes.
When HCC was compared to liver cirrhosis, chronic hepatitis, and dysplastic nodules. A significant correlation in the promoter methylation of the APC gene was observed between HCC and liver cirrhosis (OR =6.04, P<0.001), but not between HCC and chronic hepatitis, and dysplastic nodules (P>0.1). Our findings suggested that APC promoter methylation may only contribute to the progression from liver cirrhosis into HCC. Because the sample size in this study is small, more studies with large sample sizes are necessary to further validate the relationship between HCC and chronic hepatitis, and dysplastic nodules for APC promoter methylation.
Finally, the correlation of APC promoter methylation with clinicopathological features of HCC patients was also evaluated. The results revealed that APC promoter methylation was lined to HBV and HCV infection status of patients with HCC (OR =2.86, P=0.005; OR =3.50, P=0.001; respectively), but not associated with tumor stage, gender and vascular invasion status.
Substantial heterogeneity was found in cancer vs. normal live tissues and liver cirrhosis, therefore, the sensitivity analysis was performed in the current meta-analysis. Two studies (28,41) were removed in the comparison of cancer and normal live tissues, and one study (21) was removed in the comparison of cancer and liver cirrhosis. The pooled results remained significant, suggesting the stability of our analyses.
Our results compare favorably with the previous meta-analysis by Zhang et al. (56), which also found that APC promoter methylation was correlated with an increased risk of HCC in cancer vs. normal live tissues. The number of sample sizes included in the current meta-analysis (n=1,216 tissues) was larger than in the previous meta-analysis (n=944 tissues). In addition, the previous meta-analysis did not analyze whether APC promoter methylation was associated with HCC in the MSP and non-MSP subgroups.
Several limitations should be illustrated in this meta-analysis. First, our study mainly included Asian and Caucasian populations, and other ethnic groups, such as African population, were lacking. Second, based on small sample sizes, the relationship of APC promoter methylation with clinicopathological features of patients with HCC should be further studied in the future. Third, the association comparing HCC and liver cirrhosis, chronic hepatitis, or dysplastic nodules is needed in the future. Finally, based on blood samples, additional studies are essential to confirm whether APC promoter methylation may become a potential biomarker using blood samples for HCC diagnosis.
In conclusion, our findings suggest that HCC has a notably higher APC promoter methylation than normal live tissues and liver cirrhosis, but not higher than chronic hepatitis and dysplastic nodules. APC promoter methylation is correlated with HBV and HCV infection status of HCC patients, but not linked to tumor stage, gender and vascular invasion status. APC promoter methylation may be potential noninvasive biomarker in the blood. More well-matched studies with large sample sizes are essential to confirm our findings.
Full table
Acknowledgments
Funding: The research was supported by grants from the Health & Medical Science and Technology Program of Zhejiang Province (2017171691).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.71). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Suh B, Yun JM, Park S, et al. Prediction of future hepatocellular carcinoma incidence in moderate to heavy alcohol drinkers with the FIB-4 liver fibrosis index. Cancer 2015;121:3818-25. [Crossref] [PubMed]
- Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 2013;13:123-35. [Crossref] [PubMed]
- Liu Y, Chang CC, Marsh GM, et al. Population attributable risk of aflatoxin-related liver cancer: systematic review and meta-analysis. Eur J Cancer 2012;48:2125-36. [Crossref] [PubMed]
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118-27. [Crossref] [PubMed]
- Gao Y, Teschendorff AE. Epigenetic and genetic deregulation in cancer target distinct signaling pathway domains. Nucleic Acids Res 2017;45:583-96. [Crossref] [PubMed]
- Khan SA, Reddy D, Gupta S. Global histone post-translational modifications and cancer: Biomarkers for diagnosis, prognosis and treatment? World J Biol Chem 2015;6:333-45. [Crossref] [PubMed]
- Ye M, Huang T, Ying Y, et al. Detection of 14-3-3 sigma (σ) promoter methylation as a noninvasive biomarker using blood samples for breast cancer diagnosis. Oncotarget 2017;8:9230-42. [PubMed]
- Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med (Zagreb) 2015;25:161-76. [Crossref] [PubMed]
- Chen ZY, Zhang JL, Yao HX, et al. Aberrant methylation of the SPARC gene promoter and its clinical implication in gastric cancer. Sci Rep 2014;4:7035. [Crossref] [PubMed]
- Friedrich A, Kullmann F. Med Klin (Munich) 2003;98:776-82. [Familial adenomatous polyposis syndrome (FAP): pathogenesis and molecular mechanisms]. [Crossref] [PubMed]
- Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta 1997;1332:F127-47. [PubMed]
- Kámory E, Olasz J, Csuka O. Somatic APC inactivation mechanisms in sporadic colorectal cancer cases in Hungary. Pathol Oncol Res 2008;14:51-6. [Crossref] [PubMed]
- Virmani AK, Rathi A, Sathyanarayana UG, et al. Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clin Cancer Res 2001;7:1998-2004. [PubMed]
- Fodde R, Kuipers J, Rosenberg C, et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol 2001;3:433-8. [Crossref] [PubMed]
- Olkhov-Mitsel E, Zdravic D, Kron K, et al. Novel multiplex MethyLight protocol for detection of DNA methylation in patient tissues and bodily fluids. Sci Rep 2014;4:4432. [Crossref] [PubMed]
- Scarpa M, Scarpa M, Castagliuolo I, et al. Aberrant gene methylation in non-neoplastic mucosa as a predictive marker of ulcerative colitis-associated CRC. Oncotarget 2016;7:10322-31. [PubMed]
- Moelans CB, de Groot JS, Pan X, et al. Clonal intratumor heterogeneity of promoter hypermethylation in breast cancer by MS-MLPA. Mod Pathol 2014;27:869-74. [Crossref] [PubMed]
- Villanueva A, Portela A, Sayols S, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 2015;61:1945-56. [Crossref] [PubMed]
- Harder J, Opitz OG, Brabender J, et al. Quantitative promoter methylation analysis of hepatocellular carcinoma, cirrhotic and normal liver. Int J Cancer 2008;122:2800-4. [Crossref] [PubMed]
- Lee S, Lee HJ, Kim JH, et al. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol 2003;163:1371-8. [Crossref] [PubMed]
- Ding Y, Yang Q, Wang B, et al. The Correlation of MGMT Promoter Methylation and Clinicopathological Features in Gastric Cancer: A Systematic Review and Meta-Analysis. PLoS One 2016;11:e0165509 [Crossref] [PubMed]
- Xiao J, Hu CP, He BX, et al. PTEN expression is a prognostic marker for patients with non-small cell lung cancer: a systematic review and meta-analysis of the literature. Oncotarget 2016;7:57832-40. [PubMed]
- Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics 2005;21:3672-3. [Crossref] [PubMed]
- Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [Crossref] [PubMed]
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820-6. [Crossref] [PubMed]
- Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. [Crossref] [PubMed]
- Zekri AR, Bahnasy AA, Shoeab FE, et al. Methylation of multiple genes in hepatitis C virus associated hepatocellular carcinoma. J Adv Res 2014;5:27-40. [Crossref] [PubMed]
- Nishida N, Arizumi T, Takita M, et al. Quantification of tumor DNA in serum and vascular invasion of human hepatocellular carcinoma. Oncology 2013;84:82-7. [Crossref] [PubMed]
- Liu JB, Zhang YX, Zhou SH, et al. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J Gastroenterol 2011;17:4718-24. [Crossref] [PubMed]
- Huang ZH, Hu Y, Hua D, et al. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol 2011;91:702-7. [Crossref] [PubMed]
- Hua D, Hu Y, Wu YY, et al. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol 2011;91:455-60. [Crossref] [PubMed]
- Um TH, Kim H, Oh BK, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol 2011;54:939-47. [Crossref] [PubMed]
- Formeister EJ, Tsuchiya M, Fujii H, et al. Comparative analysis of promoter methylation and gene expression endpoints between tumorous and non-tumorous tissues from HCV-positive patients with hepatocellular carcinoma. Mutat Res 2010;692:26-33. [Crossref] [PubMed]
- Feng Q, Stern JE, Hawes SE, et al. DNA methylation changes in normal liver tissues and hepatocellular carcinoma with different viral infection. Exp Mol Pathol 2010;88:287-92. [Crossref] [PubMed]
- Iyer P, Zekri AR, Hung CW, et al. Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp Mol Pathol 2010;88:107-11. [Crossref] [PubMed]
- Lou C, Du Z, Yang B, et al. Aberrant DNA methylation profile of hepatocellular carcinoma and surgically resected margin. Cancer Sci 2009;100:996-1004. [Crossref] [PubMed]
- Moribe T, Iizuka N, Miura T, et al. Methylation of multiple genes as molecular markers for diagnosis of a small, well-differentiated hepatocellular carcinoma. Int J Cancer 2009;125:388-97. [Crossref] [PubMed]
- Chang H, Yi B, Li L, et al. Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp Mol Pathol 2008;85:96-100. [Crossref] [PubMed]
- Nishida N, Nagasaka T, Nishimura T, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology 2008;47:908-18. [Crossref] [PubMed]
- Nomoto S, Kinoshita T, Kato K, et al. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer 2007;97:1260-5. [Crossref] [PubMed]
- Katoh H, Shibata T, Kokubu A, et al. Epigenetic instability and chromosomal instability in hepatocellular carcinoma. Am J Pathol 2006;168:1375-84. [Crossref] [PubMed]
- Lehmann U, Berg-Ribbe I, Wingen LU, et al. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin Cancer Res 2005;11:3654-60. [Crossref] [PubMed]
- Yang B, Guo M, Herman JG, et al. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol 2003;163:1101-7. [Crossref] [PubMed]
- Yu J, Ni M, Xu J, et al. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer 2002;2:29. [Crossref] [PubMed]
- Sanchez-Carbayo M, Orenes E, Gil M, et al. Discovery of the methylation of the metastasis suppressor gene KiSS-1 in bladder cancer. J Clin Oncol 2009;27:15s, abstr 5023.
- Ullah F, Khan T, Ali N, et al. Promoter Methylation Status Modulate the Expression of Tumor Suppressor (RbL2/p130) Gene in Breast Cancer. PLoS One 2015;10:e0134687 [Crossref] [PubMed]
- Maziveyi M, Alahari SK. Breast Cancer Tumor Suppressors: A Special Emphasis on Novel Protein Nischarin. Cancer Res 2015;75:4252-9. [Crossref] [PubMed]
- Zhang W, Jiao H, Zhang X, et al. Correlation between the expression of DNMT1, and GSTP1 and APC, and the methylation status of GSTP1 and APC in association with their clinical significance in prostate cancer. Mol Med Rep 2015;12:141-6. [PubMed]
- Bilgrami SM, Qureshi SA, Pervez S, et al. Promoter hypermethylation of tumor suppressor genes correlates with tumor grade and invasiveness in patients with urothelial bladder cancer. Springerplus 2014;3:178. [Crossref] [PubMed]
- Al-Shabanah OA, Hafez MM, Hassan ZK, et al. Methylation of SFRPs and APC genes in ovarian cancer infected with high risk human papillomavirus. Asian Pac J Cancer Prev 2014;15:2719-25. [Crossref] [PubMed]
- He K, Zhang L, Long X. Quantitative assessment of the association between APC promoter methylation and breast cancer. Oncotarget 2016;7:37920-30. [PubMed]
- Shen C, Sheng Q, Zhang X, et al. Hypermethylated APC in serous carcinoma based on a meta-analysis of ovarian cancer. J Ovarian Res 2016;9:60. [Crossref] [PubMed]
- Huang T, Li J, Zhang C, et al. Distinguishing Lung Adenocarcinoma from Lung Squamous Cell Carcinoma by Two Hypomethylated and Three Hypermethylated Genes: A Meta-Analysis. PLoS One 2016;11:e0149088 [Crossref] [PubMed]
- Pang JM, Deb S, Takano EA, et al. Methylation profiling of ductal carcinoma in situ and its relationship to histopathological features. Breast Cancer Res 2014;16:423. [Crossref] [PubMed]
- Zhang C, Li J, Huang T, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget 2016;7:81255-67. [PubMed]