
IL-10 and IL-10RB gene polymorphisms are correlated with hepatitis B-related hepatocellular carcinoma in the Chinese Han population
Introduction
Hepatocellular carcinoma (HCC) is the third major fatal cause of cancer globally, and the most common etiology for this cancer is chronic infection with hepatitis B virus (HBV) (1,2). China has the largest disease burdens of HBV infection and HCC globally (3). However, the precise mechanism underlying HCC remains poorly understood. Cytokines are key immunomodulatory molecules in the immune response system and are reported to be important factors in HCC progression. It has been reported that cytokine genotypes may be critical to the genetic regulation of immune responses to HBV infection and in determining an individual’s level of HCC risk. Numerous studies have demonstrated that single nucleotide polymorphisms (SNPs) appear to be consistently associated with HCC risk (4). Multiple SNPs in cytokine genes and their receptors are known to influence different clinical outcomes of HBV-infected patients and to be involved in the molecular mechanisms of hepatitis-related HCC (5-7).
Interleukin-10 (IL-10) is a powerful multifunctional cytokine produced by T-helper 2 cells and macrophages that exhibits complex biological activity. The IL-10 gene, which is located in 1q31-32, is composed of 5 exons and 4 introns (8). In recent years, SNPs in IL-10 and IL-10 receptor beta (IL-10RB) have been shown to be closely correlated to the pathogenesis of many cancers (9-11). The IL-10RB, interferon-α (IFN-α) and the interferon-γ (IFN-γ) receptors are located in a cluster of class II cytokine receptor gene at 21q22 (12). One IL-10RB gene polymorphism (IL10RB E47K) has been associated with a risk of HBV persistence (13). The relationships between IL-10 and IL-10RB gene polymorphisms and inflammation, such as chronic hepatitis B (CHB) and HCC, have aroused widespread interest.
In view of this and based on SNP genotype data in the HapMap phase II + phase III database, four tagSNPs (rs3790622, rs1518110 and rs1800872 in the IL-10 gene and rs2244305 in the IL-10RB gene) were selected for genotyping because they cover about 80% of the SNP sites (MAF >0.1). To examin the correlations between the four SNPs and susceptibility to HCC and CHB, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) were applied to analyze IL-10 and IL-10RB gene polymorphisms in HCC, CHB and healthy groups. In this study, we present data describing a relationship between IL-10 and IL-10RB gene polymorphisms and susceptibility to hepatitis B-related HCC.
Methods
Study population
Peripheral blood samples were collected from 430 HCC patients, 342 CHB patients and 371 healthy donors of the Chinese Han population from November 2001 to April 2010. HCC patients were recruited from Peking University People’s Hospital (Beijing, China), the Affiliated Hospital of Guilin Medical University (Guilin, China) and Henan Cancer Hospital (Henan, China). The diagnosis of HCC was according to imaging and presence of alpha-fetoprotein (AFP) level >400 ng/mL. In cases where AFP <400 ng/mL, the diagnosis of HCC was made by having two typical dynamic imaging studies or one typical imaging study in patients with cirrhosis and a nodule >1 or 2 cm, as per the Asia-Pacific Association of the Study of the Liver (APASL) guidelines. CHB donors were patients diagnosed at Peking University People’s Hospital who met the diagnostic standard (14). Healthy donors were selected randomly from individuals conducting the medical examination in Peking University People’s Hospital. Donors with HBV or HCV infection, liver cirrhosis and cancer were excluded. The detailed clinical and biological characteristics of all patients are showed in Table 1. The blood samples were collected and rapidly transferred to −80 °C for storage. The Center Ethics Committees of Henan Cancer Hospital authorized this work. Written informed consent based on the Declaration of Helsinki was obtained from all patients.

Full table
SNP genotyping
From the SNP genotypes of the IL-10 and IL-10RB genes available in the Hapmap phase II + phase III database, four tagSNPs (rs3790622, rs1518110 and rs1800872 in the IL-10 gene and rs2244305 in the IL-10RB gene) were selected based on haplotype analysis using Haploview 4.2 with the following filtering criteria: r2>0.8 and a minimum allele frequency (MAF) >0.1. These four SNPs covered nearly 80% of the MAF >0.1 SNPs.
DNA was extracted from peripheral blood leukocytes using a standard phenol-chloroform method. Polymorphism analysis was conducted using the high-throughput array platform based on the MassARRAY molecular weight (Sequenom Corporation, CA, USA) according to the manufacturer’s protocol, as previously described (15). First, PCR and single-base extension primers were designed by Sequenom Assay Design 3.1 software (Table 2). Second, target fragments were amplified from the DNA samples using a semi-nested PCR reaction in a 384-well SpectroCHIP microarray. Finally, after hybridization of the SNP sites with the probes, the molecular differences in bases between the extension products and the non-extension primers were detected by an MALDI-TOF Mass Spectrometer. The Sequenom MassARRAY RT software was used to analyze the result.

Full table
Statistical analysis
Statistical analysis was performed using SPSS software (version 13.0, SPSS Inc., Chicago, IL, USA). The allele and genotype frequencies of the IL-10 and IL-10RB genes were calculated by direct counting. The chi-square test was used to compare the allele and genotype frequencies between the groups, and the Hardy-Weinberg equilibrium was used to determine the genotype distribution. Odds ratio (OR) and 95% confidence intervals (95% CI) were calculated using logistic regression. Data were considered statistically significant at P<0.05.
Results
The clinical characteristics of all donors
No significant differences were observed in the age or the aspartate aminotransferase (AST), alanine aminotransferase (ALT), γ-glutamyl transpeptidase (GGT) or total bilirubin (TBIL) levels of the HCC or CHB groups when compared to the healthy group (all P>0.05, Table 1). All healthy donors were HBsAg negative, but 92.6% of the HCC patients were HBsAg positive; all donors were anti-HCV negative. The AFP level and the frequency of cirrhosis in the HCC group were significantly higher than those of CHB group. The distributions of gender and alcohol abuse showed that male, alcohol-abusing donors accounted for the majority of the HCC, CHB and healthy donors. All donors were randomized into three groups; thus, these features represented a true natural history of the incidence of CHB and HCC in Han population.
The allele frequency distribution of the IL-10 and IL-10RB gene polymorphisms in all donors
As shown in Table 3, for rs3790622 in IL-10, the differences in the C and T allele frequencies between the HCC and CHB groups and between the HCC and healthy groups were not significant (P=0.41 and P=0.25, respectively). However, T allele frequency at rs1518110 in IL-10 (OR =1.32, 95% CI: 1.07–1.63, P=0.01), A allele frequency at rs1800872 in IL-10 (OR =1.33, 95% CI: 1.08–1.65, P<0.01), and C allele frequency at rs2244305 in IL-10RB (OR =1.44, 95% CI: 1.14–1.82, P<0.01) were higher in HCC patients than in the healthy controls. Compared to CHB patients, the above three SNP allele frequencies were also significantly higher in HCC patients. No significant differences were found in the allele frequencies at rs3790622, rs1518110, rs1800872 and rs2244305 between the CHB and healthy control groups (P=0.77, P=0.78, P=0.62, and P=0.68, respectively).
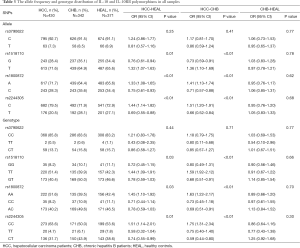
Full table
The genotype distribution of the IL-10 and IL-10RB gene polymorphisms in all donors
The genotype frequencies of the IL-10 gene polymorphism at rs3790622 between the HCC and CHB groups and between the HCC and healthy groups had no significant differences (P=0.71 and P=0.44, respectively, Table 3). Compared to healthy controls, the distribution frequency of the TT genotype at rs1518110 was significantly higher (OR =1.44, 95% CI: 1.09–1.91; P=0.03) in HCC patients; in HCC patients, the respective distribution frequencies of the TT, GT and GG genotypes at rs1518110 were 51.4%, 40.4% and 8.2%, and were comparable to the frequencies of 42.3%, 46.6% and 11.1% for healthy controls. The distribution frequency of the AA genotype at rs1800872 was significantly higher (OR =1.45, 95% CI: 1.10–1.92; P=0.03) in HCC group; in HCC patients, the respective distribution frequencies of the AA, AC and CC genotypes for SNP rs1800872 were 51.6%, 40.2% and 8.2%, which were comparable to the frequencies of 42.4%, 46.5% and 11.1% in the healthy controls. The distribution frequency of the CC genotype (rs2244305) was significantly higher (OR =1.51, 95% CI: 1.14–2.01; P=0.01) in HCC patients compared to healthy controls; in HCC patients, the respective distribution frequencies of the CC, CT and TT genotypes of SNP rs2244305 were 63.6%, 31.7% and 4.7%, which were comparable to the frequencies of 53.6%, 38.5% and 7.8% for the control group. Compared to the CHB group, the HCC patients also showed higher frequencies of TT genotype at rs1518110 (OR =1.59, 95% CI: 1.92–2.12, P<0.01), AA genotype at rs1800872 (OR =1.63, 95% CI: 1.22–2.17, P<0.01) and CC genotype at rs2244305 (OR =1.75, 95% CI: 1.31–2.34, P<0.01). However, differences in genotype frequency at rs3790622, rs1518110, rs1800872 and rs2244305 were not observed between the CHB patients and the healthy controls (P=0.77, P=0.66, P=0.70, and P=0.30, respectively).
The stratified analysis of the association between the IL-10 and IL-10RB genotypes and HCC clinical pathology characteristics
Considering the genotypes or alleles of rs1518110, rs1800872 and rs2244305 were significantly correlated with HCC susceptibility, we further analyzed the genotypic effects of the four SNPs in 234 HCC cases with detailed clinical information, including age, gender, history of alcohol abuse and smoking, tumor size, tumor number, Barcelona Clinic Liver Cancer (BCLC) stage, the degree of hepatic cirrhosis, distant metastasis, lymph node metastasis and recurrence (Table 4). In the stratified analysis, we observed significant correlations between the GG/GT genotypes at rs1518110 and the CC/AC genotypes at rs1800872 with individual smoking habit and HCC risk (P<0.001) and with an increased rate of tumor recurrence in HCC patients (P<0.01). In HCC patients, the CC genotype of rs2244305 was significantly correlated with multiple tumor number, B or C BCLC stages, distant metastasis and lymph node metastasis (P<0.01, P<0.01, P=0.01, and P=0.02, respectively).
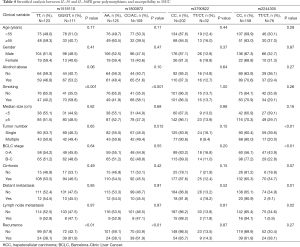
Full table
Discussion
IL-10 and IL-10RB are regarded as important anti-inflammatory cytokines in humans and are significant for maintaining immune system balance by down-regulating the pro-inflammatory response (16). IL-10 potentially has dual biological functions in the promotion and inhibition of cancer and may regulate tumor susceptibility and development (17). Studies of IL-10 gene polymorphisms revealed that IL-10 SNPs are related to cancer susceptibility and affect the severity and progress of the disease as well as the IL-10 expression level (7). Several studies have investigated the associations of polymorphisms in IL-10 and its receptor IL-10RB with cancer susceptibility and prognosis (11). Recently, a correlation between polymorphisms in the IL-10 gene promoter region and a variety of tumors, including HCC, has been reported (18,19).
In our study, compared to the healthy control and CHB groups, the T allele frequency of rs1518110, the A allele (rs1800872) and C allele (rs2244305) have a significantly higher frequency in HCC patients. The distribution frequencies of the TT genotype in SNP rs1518110, the AA genotype in SNP rs1800872 and the CC genotype in SNP rs2244305 were higher in HCC patients than in CHB patients. It was hypothesized that patients with the IL-10 rs1518110, rs1800872 and rs2244305 polymorphisms were possibly susceptible to HCC. However, the allele and genotype frequencies of the IL-10 gene SNP rs3790622 among the HCC, healthy control and CHB patients showed no significant difference. Therefore, it is supposed that IL-10 gene SNP rs3790622 may not be associated with the outcome of hepatitis B-related HCC in the Chinese Han population. In the previous studies, IL-10 gene polymorphisms were important for HBV infection-induced immune response. The IL-10/-592 C/C and IL-10/1927 A/A genotypes are related to a higher risk of HCC (20). IL-10 haplotype 2 (IL10-ht2) significantly up-regulated the risk of developing cirrhosis and HCC in chronic HBV-infected patients (21). Our results also showed that the allele and genotype frequencies of four SNPs (rs3790622, rs1518110 and rs1800872 in the IL-10 gene and rs2244305 in the IL-10RB gene) in CHB group were not different from those of healthy control groups. This is compatible with a previous study that IL-10 promoter polymorphisms were uncorrelated with HBV infection prognosis (22). It is, therefore, clear that IL-10 gene SNPs are strongly associated with HCC.
Our findings also showed that significant interactions between the IL-10 gene genotypes (GG/GT genotypes at rs1518110, CC/AC genotypes at rs1800872) and smoking might increase the risk of HCC and the rate of tumor recurrence in HCC patients. Inversely, the TT genotype (rs1518110) and AA genotype (rs1800872) are possibly protective genotypes for HCC recurrence. Interestingly, this finding was consistent with a previous report that genotypes at rs1800872 in IL-10 were linked to a high risk of gastric cancer in smokers (23). Moreover, the CC genotype at rs2244305 was significantly associated with tumor number, BCLC stage, and distant and lymph node metastasis in HCC patients. Inversely, the TT/CT genotypes at rs2244305 are possibly protective genotypes for tumor metastasis in HCC patients.
The allele and genotype frequencies of rs1518110, rs1800872 and rs2244305 in CHB patients and healthy controls have no difference. However, the allele and genotype frequencies of these SNPs were significantly different between the HCC patients and the healthy controls as well as the CHB group. It is supposed that mutations at the SNP sites can affect IL-10 and IL-10RB protein expression, T-helper 1/2 cell differentiation and the balance of immune mechanisms. This suggests that IL-10 and IL-10RB may play a role in inhibiting anti-inflammatory cytokine reduction, delaying HBV-related inflammatory responses and inducing HCC, which may be caused by a cumulative effect of these processes.
Several studies have shown the central role of inflammatory cytokines such as IL10 in the continuous and progressive immune response that eventually leads to CHB, hepatitis B cirrhosis (HBC) and HCC during HBV infection. This study provides further evidence that functional IL-10 (rs1518110 and rs1800872) and IL-10RB (rs2244305) polymorphisms may promote the risk of hepatitis B-related HCC. However, a moderate sample size limits the statistical power of our study. We will further determine our findings and clarify the hepatitis B-related HCC genetic mechanism. Therefore, more in-depth investigations of the relationships between IL-10 and IL-10RB gene polymorphisms and host genetic susceptibility to hepatitis B-related HCC should be conducted to provide additional clues toward understanding the occurrence and development of hepatitis B-related HCC.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81001072 and U1304818 to H Zhang), the Beijing Natural Science Foundation (7152151 to H Zhang), the Key Project of Science and Technology of Henan Province (122102310056 to J Zhou), the Project of Science and Technology of Henan Province (Z2013912078 to J Zhou), and the Jiangsu Key Laboratory of Medicine Science and Laboratory Medicine (JSKLM-2014-008 to H Zhang).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.04.09). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by The Center Ethics Committees of Henan Cancer Hospital and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55. [Crossref] [PubMed]
- Byam J, Renz J, Millis JM. Liver transplantation for hepatocellular carcinoma. Hepatobiliary Surg Nutr 2013;2:22-30. [PubMed]
- Tanaka M, Katayama F, Kato H, et al. Hepatitis B and C Virus Infection and Hepatocellular Carcinoma in China: A Review of Epidemiology and Control Measures. J Epidemiol 2011;21:401-16. [Crossref] [PubMed]
- Gonzalez SA. Novel biomarkers for hepatocellular carcinoma surveillance: has the future arrived? Hepatobiliary Surg Nutr 2014;3:410-4. [PubMed]
- Nieters A, Yuan JM, Sun CL, et al. Effect of cytokine genotypes on the hepatitis B virus-hepatocellular carcinoma association. Cancer 2005;103:740-48. [Crossref] [PubMed]
- Dondeti MF, El-Maadawy EA, Talaat RM. Hepatitis-related hepatocellular carcinoma: Insights into cytokine gene polymorphisms. World J Gastroenterol 2016;22:6800-16. [Crossref] [PubMed]
- Bidwell J, Keen L, Gallagher G, et al. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun 1999;1:3-19. [Crossref] [PubMed]
- Eskdale J, Kube D, Tesch H, et al. Mapping of the human IL10 gene and further characterization of the 5' flanking sequence. Immunogenetics 1997;46:120-8. [Crossref] [PubMed]
- Ni J, Ye Y, Teng F, et al. Interleukin 10 polymorphisms and cervical cancer risk: a meta-analysis. Int J Gynecol Cancer 2013;23:126-33. [Crossref] [PubMed]
- Yu KD, Chen AX, Yang C, et al. The associations between two polymorphisms in the interleukin-10 gene promoter and breast cancer risk. Breast Cancer Res Treat 2012;131:27-31. [Crossref] [PubMed]
- Kasamatsu T, Saitoh T, Ino R, et al. Polymorphism of IL-10 receptor beta affects the prognosis of multiple myeloma patients treated with thalidomide and/or bortezomib. Hematol Oncol 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Hikami K, Ehara Y, Hasegawa M, et al. Association of IL-10 receptor 2 (IL10RB) SNP with systemic sclerosis. Biochem Biophys Res Commun 2008;373:403-7. [Crossref] [PubMed]
- Frodsham AJ, Zhang L, Dumpis U, et al. Class II cytokine receptor gene cluster is a major locus for hepatitis B persistence. Proc Natl Acad Sci U S A 2006;103:9148-53. [Crossref] [PubMed]
- Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology 2009;50:661-2. [Crossref] [PubMed]
- Chen Y, Zhang H, Liao W, et al. FOXP3 gene polymorphism is associated with hepatitis B-related hepatocellular carcinoma in China. J Exp Clin Cancer Res 2013;32:39. [Crossref] [PubMed]
- Glocker EO, Kotlarz D, Klein C, et al. IL-10 and IL-10 receptor defects in humans. Ann N Y Acad Sci 2011;1246:102-7. [Crossref] [PubMed]
- Villalta SA, Rinaldi C, Deng B, et al. Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum Mol Genet 2011;20:790-805. [Crossref] [PubMed]
- Yang Y, Luo C, Feng R, et al. The TNF-alpha, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol 2011;137:947-52. [Crossref] [PubMed]
- Migita K, Miyazoe S, Maeda Y, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection-association between TGF-β1 polymorphisms and hepatocellular carcinoma. J Hepatol 2005;42:505-10. [Crossref] [PubMed]
- Tseng LH, Lin MT, Shau WY, et al. Correlation of interleukin-10 gene haplotype with hepatocellular carcinoma in Taiwan. Tissue Antigens 2006;67:127-33. [Crossref] [PubMed]
- Shin HD, Park BL, Kim LH, et al. Interleukin 10 haplotype associated with increased risk of hepatocellular carcinoma. Hum Mol Genet 2003;12:901-6. [Crossref] [PubMed]
- Sofian M, Kalantar E, Aghakhani A, et al. No Correlation Between Interleukin-10 Gene Promoter Polymorphisms and Hepatitis B Virus Infection Outcome. Hepat Mon 2013;13:e8803 [Crossref] [PubMed]
- Kim J, Cho YA, Choi IJ, et al. Effects of interleukin-10 polymorphisms, Helicobacter pylori infection, and smoking on the risk of noncardia gastric cancer. PLoS One 2012;7:e29643 [Crossref] [PubMed]


