
LAPTM4B-35 expression was correlated with favorable post-operation survival in pancreatic cancer patients
Introduction
Pancreatic cancer (PC) is notorious for its aggressive and malignant biological behaviors and is the fourth leading cause of cancer related death (1). Most PC patients cannot be diagnosed at early stage because their symptoms are not obvious, and a large amount of patients are not suitable for surgical resection due to its severe invasion and metastasis. For the patients who receive surgical resection, 5-year survival rate is less than 20% and 10-year survival rate even decreases to approximately 2% (2). So, it will be quite helpful if we can diagnose this disease as early as possible and elucidate the mechanisms underlying its invasive behaviors.
Lysosome-associated protein transmembrane 4 beta (LAPTM4B) was first cloned in hepatocellular carcinoma (HCC). LAPTM4B gene is located at chromosome 8q22.1 and it contains seven exons and six introns (3). LAPTM4B gene encodes a member of the mammalian 4-tetransmembrane spanning protein superfamily. LAPTM4B-24 and LAPTM4B-35 are two proteins with different molecular weight encoded by LAPTM4B gene (4). LAPTM4B-35 has been found to be overexpressed in a variety of solid tumors compared with LAPTM4B-24, such as HCC (5-7), breast cancer (8), ovarian cancer (9), gastric cancer (10,11), cervical cancer (12), colorectal cancer (13) and so on, wherein LAPTM4B-35 was also found to be correlated with invasive behaviors of these cancers.
However, few studies have focused on the relationship between LAPTM4B and PC. Our study aimed to evaluate the expression of LAPTM4B-35 in tumor tissues and paired non-cancerous tissues in PC patients and to assess the correlation of LAPTM4B-35 expression with clinicopathological features or post-operation survival of PC patients, which may be helpful to elucidate the relationship between LAPTM4B-35 expression and PC.
Methods
Patients and samples
Tumor tissues and paired non-cancerous tissues were obtained from ninety-three patients who were diagnosed as PC and received surgical resection during 2008–2014 at Peking University Cancer Hospital, Beijing, China. Tumor tissues and paired non-cancerous tissues were formalin-fixed immediately after resection and then embedded with paraffin. All patients had clinical records and none of them received chemotherapy or radiation therapy before surgery. These patients were followed up for at least 5 years after surgery. The detailed clinicopathological features of these 93 PC patients were described in Table 1. This study was approved by the Ethical Committee of Peking University Cancer Hospital (Ethical approval number: 2016KT41), and written informed consent was acquired from each patient prior to initiation.
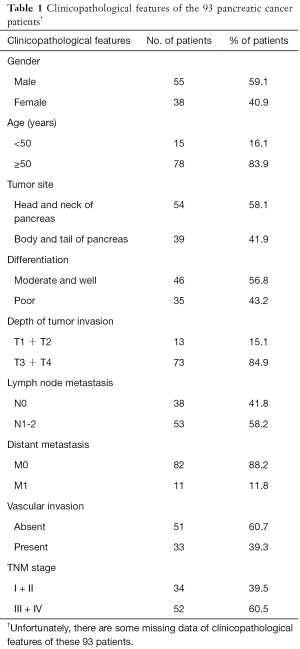
Full table
Immunohistochemistry
The 4 µm thick tissue sections were baked at 72 °C for 1 hour, and then dewaxed in xylene and rehydrated in graded alcohol concentrations. The 3% hydrogen peroxide was used to block the activity of endogenous peroxidase for 15 minutes. After PBS washing 3 times, the antigen retrieval was performed by heating the sections in a microwave for 10 min in 0.01 mol/L citrate buffer (pH =6.0). When cooling to room temperature, the sections were blocked by goat serum containing 5% BSA (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China) for 1 hour at room temperature. Then the sections were incubated with LAPTM4B-35 antibody (gifted by Pro. RL Zhou, Department of Cell Biology, School of Basic Medical Sciences, Peking University, Beijing, China) at a dilution of 1:1,500 at 4 °C overnight. The following day, the ready-to-use EnVisionTM regent (EnVisionTM detection system peroxidase/DAB, rabbit/mouse; Dako, Glostrup, Denmark) was used to bind the primary antibody. The 3,3'-diaminobenzidine tetrahydrochloride (Dako, Glostrup, Denmark) was used to visualize the reaction. Then the sections were counterstained with hematoxylin and dehydration was performed in the sequence of graded alcohol concentrations and xylene.
Evaluation of staining
LAPTM4B-35 expression levels were classified semi-quantitatively based on the total combined scores of positive-staining tumor cell percentage and staining intensity by two independent pathologists who were blind to all patients’ clinical data. The percentage of positive cells (PP) was scored as 0 (negative), 1 (<25%), 2 (25–75%), and 3 (>75%) respectively while the staining intensity (SI) was scored as 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The immunoreactivity score (IRS) was defined as the sum of PP and SI, and IRS <1 was regarded as “negative” while IRS ≥1 was “positive”.
Cell lines and cell culture
Five PC cell lines (SW1990, PANC-1, CFPAC-1, AsPC-1 and BxPC-3) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GibcoBRL, Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS), 100 µg/mL penicillin and 100 µg/mL streptomycin. And all cell lines were maintained at 37 °C in a humid condition with 5% CO2.
Isolation of total RNA, reverse transcription-polymerase chain reaction (RT-PCR) and quantitative real-time PCR
We explored LAPTM4B-35 expression of mRNA level by means of RT-PCR and quantitative real-time PCR.
Total RNA of cell lines was isolated using Trizol reagent (Invitrogen, Life Technologies, Carlsbad, California, USA) while total RNA of 13 frozen tissues and paired non-cancerous tissues were isolated using RNeasy Mini Kit (Cot. 74104, QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. cDNA was synthesized from the 1 µg total RNA in a 20 µL reaction volume by RT using 5× All-In One RT MasterMix (Abm, Canada) with a procedure of 25 °C for 10 minutes, 42 °C for 50 minutes and 85 °C for 5 minutes.
RT-PCR amplification was performed using the following LAPTM4B-35 and beta-actin gene specific primers: LAPTM4B-35, 5'-CAAGAATTTCCCCCACAACATC-3' (sense) and 5'-CGGCCTGGTGCTTTCTAATG-3' (antisense); and beta-actin, 5'-TTAGTTGCGTTACACCCTTTC-3' (sense) and 5'-ACCTTCACCGTTCCAGTTT-3' (antisense). Beta-actin was used to normalize LAPTM4B-35 gene expression. The PCR procedure was as follows: pre-denaturation at 94 °C for 5 minutes, 35 PCR cycles and a final elongation at 72 °C for 10 minutes. Each PCR cycle contained denaturation at 94 °C for 45 seconds, annealing at 60 °C for 45 seconds and extension at 72 °C for 45 seconds. PCR products were separated by gel electrophoresis on 2% agarose gel stained with GelRed (Biotium, USA). The band intensity was measured on an UVP EC3 Imaging System (Uvp Inc., Upland, CA, USA).
When performing real-time PCR, we used expressed Alu repeats (EAR) (14) as loading control instead of beta-actin with primer sequences of 5'-GAGGCTGAGGCAGGAGAATCG-3' (sense) and 5'-GTCGCCCAGGCTGGAGTG-3' (antisense). And we performed EAR amplification using cDNA diluted 50 times. Real-time PCR was performed with pre-denaturation at 95 °C for 10 minutes, 40 cycles of denaturation at 95 °C for 15 seconds, annealing at 60 °C for 1 minutes and extension at 72 °C for 30 seconds. All reactions were performed in triplicate. The LAPTM4B-35 expression was measured using 2−ΔΔCT method.
Western blot analysis
Total protein was extracted from each cell line by 1× sodium dodecyl sulfate (SDS) lysis buffer. After denaturation at 95 °C for 5 minutes, equal amounts of protein (50 µg) were separated by 8% SDS-PAGE gel. After electrophoresis, the proteins were transferred onto polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with 5% skim milk for 2 hours at room temperature and then incubated by rabbit anti-human LAPTM4B-35 antibody (1:800) and mouse anti-human beta-actin antibody (1:20,000, sigma, Cot. A4552) at 4 °C overnight. Beta-actin was used as a loading control. After being washed 3 times by PBST (phosphate-buffered saline with 0.1% Tween-20) buffer, the membranes were incubated with secondary horseradish peroxidase conjugated anti-rabbit and anti-mouse antibodies respectively at a dilution of 1:5,000 at room temperature for 2 hours. Finally, protein bands were evaluated with an enhanced chemiluminescence system.
Statistical analysis
The statistical analyses were performed using SPSS software, version 13.0 (SPSS, Chicago, IL, USA). Two-tailed chi-squared test (χ2) or Fisher’s exact test was used to compare LAPTM4B-35 protein expression between PC tissues and paired non-cancerous tissues, and to evaluate the relationship between LAPTM4B-35 expression and clinicopathological factors. The differential expression of LAPTM4B-35 mRNA between tumor tissues and paired non-cancerous tissues was compared by using a nonparametric test. Kaplan-Meier survival analysis was used to evaluate patients’ post-operation survival and P values were calculated by the log rank test. Multivariate survival analysis was performed with Cox proportional hazards regression model to identify the independent parameters affecting overall survival. In all cases, a two-sided P value less than 0.05 was considered statistically significant.
Results
LAPTM4B-35 RNA and protein expression in pancreatic cell lines
We first examined RNA expression of LAPTM4B-35 gene by RT-PCR in five pancreatic cell lines (SW1990, PANC-1, CFPAC-1, AsPC-1 and BxPC-3). The result showed that LAPTM4B-35 gene was expressed at different levels in these cell lines, wherein LAPTM4B-35 was found highly expressed in PANC-1, CFPAC-1 and BxPC-3 compared with SW1990 and AsPC-1 (shown in Figure 1A).

Furthermore, we evaluated the expression of LAPTM4B-35 protein in 5 PC cell lines by western blotting assay and found that LAPTM4B-35 protein was expressed at different levels (shown in Figure 1B). We also noted that expression level of LAPTM4B-35 protein was different from RNA expression in these cell lines, that is, LAPTM4B-35 protein was highly expressed in SW1990, CFPAC-1 and AsPC-1. The difference between mRNA and protein expression might mainly be attributed to translational modification.
LAPTM4B-35 RNA and protein expression in pancreatic tissues
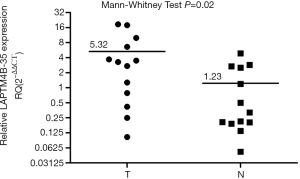
We assessed RNA expression of LAPTM4B-35 gene in 13 pairs of primary PC tissues and paired non-cancerous tissues by quantitative real-time PCR. As shown in Figure 2, we found that the mRNA expression level of LAPTM4B-35 was higher in tumor tissues than in pancreatic normal tissues (P=0.02).
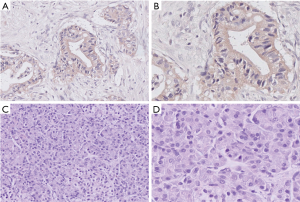
We also examined expression of LAPTM4B-35 protein by immunohistochemistry assay in tumor tissue and paired non-cancerous tissues from 93 PC patients. In these specimens, LAPTM4B-35 was expressed in 60 of 93 (64.5%) tumor tissues and 7 of 93 (7.5%) paired non-cancerous tissues. And LAPTM4B-35 protein was predominantly localized in the cytoplasm of PC cells. The expression of LAPTM4B-35 in tumor tissues was markedly higher than in non-cancerous tissues (P<0.001). Representative images of LAPTM4B-35 expression are shown in Figure 3.
Correlation of LAPTM4B-35 expression with clinicopathological parameters in PC patients
Next, we assessed the relationship between LAPTM4B-35 protein expression and clinicopathological features of PC. As shown in Table 2, we found that LAPTM4B-35 expression was correlated with tumor differentiation (P=0.039). Meanwhile, there was a tendency that LAPTM4B-35 expression was a little higher in patients with more favorable clinicopathological features, including moderate and well differentiation, weaker tumor invasion, no lymph node metastasis, no distant metastasis, absence of vascular invasion and early TNM stage though without statistical difference (shown in Figure 4).
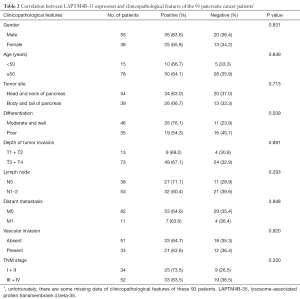
Full table

Correlation of LAPTM4B-35 protein expression with post-operation survival of PC patients
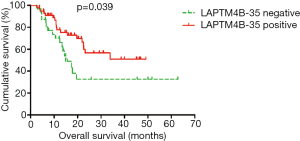
The follow-up time was measured from the operation date to the time of death or the last follow-up visit, which was from May 2008 to December 2015. During the follow-up period, 37 (39.8%) patients died of PC. Median survival time was 14.2 months (range from 2.5 to 62.7 months). The median survival time was 17.64±16.04 months for LAPTM4B-35 negative patients and 19.2±13.38 months for LAPTM4B-35 positive patients, respectively. We used Kaplan-Meier survival analysis and log rank test to evaluate the relationship between LAPTM4B-35 expression or clinicopathological features and post-operation survival of PC patients. The survival curves showed that patients with positive LAPTM4B-35 expression had a better post-operation survival than those with negative LAPTM4B-35 expression (P=0.039; Figure 5).
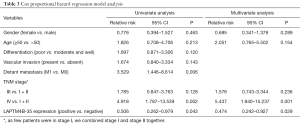
Various clinicopathological features that might affect overall survival were evaluated by univariate survival analysis, and we found that distant metastasis (P=0.006), advanced TNM stage (IV vs. I + II, P=0.002) and negative LAPTM4B-35 expression (P=0.043) were related to poor post-operation survival of PC patients. And multivariate Cox regression survival analysis further showed that LAPTM4B-35 (positive vs. negative, P=0.029, OR =0.474, 95% CI: 0.242–0.927) and TNM stage (IV vs. I + II, P=0.001, OR =5.437, 95% CI: 1.940–15.237) were independent factors for post-operation survival of PC patients. For detailed information, please see Table 3.
Full table
Discussion
PC is a kind of aggressive carcinoma especially at its advanced stage and it is one of the most lethal human tumors. A study relating to familial PC has shown that PC might be quite heterogeneous with numerous gene deletions, amplifications and so on (15). Therefore, the development and progression of PC may involve various factors. It is important to find out markers to assist diagnosis, to monitor progression or to predict prognosis of PC patients.
Lysosome-associated protein transmembrane 4 beta (LAPTM4B) is a kind of transmembrane oncoprotein that belongs to the mammalian LAPTM family. Overexpression of LAPTM4B has been proved in several kinds of solid tumors and has been related with malignant behaviors of these tumors. Furthermore, LAPTM4B was found to be an independent prognosis factor in several kinds of tumors (5,7-13).
While, the mechanism of LAPTM4B in tumor development and progression remains elusive. Overexpression of LAPTM4B-35 can promote cell proliferation, cell migration and invasion in HCC through upregulating proliferation-promoting transcription factors such as c-Myc (16). In NSCLC, LAPTM4B increased NRF2 target genes such as heme oxygenase 1 (HMOX1) and the LAPTM4B/NRF2 signaling axis modulated differentially in different cell lines with distinct mutational, genetic and epigenetic profiles and was being more activated in KRAS mutant lung cancer cells (17). And LAPTM4B can play its role with other biomarkers. Overexpression of LAPTM4B-35 in combination with positive argininosuccinate synthetase (ASS) expression was a negative prognostic marker for HCC patients (18), and combined overexpression of LAPTM4B-35 and VEGF was an independent factor for both OS and DFS in cervical carcinoma patients (12).
LAPTM4B is also related with multidrug resistance. Upregulation of LAPTM4B-35 can motivate multidrug resistance by interaction with multidrug resistance 1 (MDR1) and activation of PI3K/AKT signaling pathway (19).
Recently, Klionsky et al. have found that LAPTM4B can play a role in autophagy. In lung cancer cells, suppression of LAPTM4B can cause an increase in LC3-II and p62 protein levels, which suggests decreased autophagosome turnover and fusion with lysosomes as well as abnormal autophagic flux. In addition, in serum deprived lung cancer cells, LAPTM4B co-localized with LC3. LAPTM4B was an important mediator of autophagy for survival of lung cancer cells with serum starvation (17).
Few studies have focused on the expression of LAPTM4B and its function in PC, so the present study aimed to assess the relationship between LAPTM4B-35 expression and PC. In our study, we have detected high LAPTM4B-35 protein expression in one primary PDAC cell lines (BxPC-3) and two metastatic PDAC cell lines (SW1990 and CFPAC-1). On the other hand, LAPTM4B-35 protein expression was significantly lower in PANC-1 (a primary PDAC cell line) and AsPC-1 (a metastatic PDAC cell line). From these findings, we cannot see a regular tendency, which may be attributed to multiple mutations during cellular immortalization. Even so, these cell lines can provide useful in vitro tools which can be manipulated for over-expression or knockdown so as to carry out subsequent functional or mechanism studies. We also noted that expression level of LAPTM4B-35 protein was different from RNA expression in these cell lines, and the difference might be attributed to translational modification and it may also be caused by regulation of upstream factors which might influence protein expression.
We also have found that expression of LAPTM4B-35 was higher in tumor tissues than paired non-cancerous tissues of PC patients. And there was a tendency that expression of LAPTM4B-35 was elevated in tumors with favorable clinicopathological features even though there was no statistical significance. LAPTM4B-35 was highly expressed in PC with moderate and well differentiation, relatively superficial depth of tumor invasion, no lymph node metastasis, no distant metastasis, absence of vascular invasion and early TNM stage. LAPTM4B-35 expression showed a gradually decreasing tendency with tumor progression, which has not been reported before.
Moreover, survival analysis showed that patients with positive LAPTM4B-35 expression had a better post-operation survival than those with negative LAPTM4B-35 expression, which was opposite to Dr. Zhou’s research before (20). This difference might be due to the following reasons. Firstly, in Dr. Zhou’s paper, LAPTM4B-35 protein was classified as low expression and high expression, while in our research we classified its expression as negative and positive. Different methods of identification may possibly cause different results. Secondly, it has been reported that LAPTM4B can play a dual function during tumor progression. The N-terminal part of LAPTM4B might be involved in signal transduction thus enhancing cell proliferation whereas the C-terminus might be responsible for the correct protein sorting to late endosomes and lysosomes (21). Thirdly, the different findings in the two studies might be caused by relatively small sample size and relatively short follow-up time. And in multivariate survival analysis, LAPTM4B-35 and TNM stage were independent factors for post-operation survival of PC patients. So, in clinical practice, we can evaluate the expression of LAPTM4B-35 in tumor specimen after surgery. If its expression is positive, then it might imply a favorable outcome, but we must combine routine postoperative re-examination to monitor the progression of PC patients. And in future we will expand the sample size and improve the post-operation follow-up to further validate our results.
Therefore, we inferred that LAPTM4B-35 might be up-regulated at the development stage of PC and doesn’t participate much during the progression of this disease, which was not the same with other solid tumors (5,8,11). And this needs functional and mechanism validation. In Professor Zhou’s lab, they have found that LAPTM4B-35 can be detected through enzyme-linked immunosorbent assay (ELISA) as exosome form in serum of HCC patients (unpublished data), so in future we will try to assess the LAPTM4B-35 level in PC patients, so as to assist diagnosis by combing other serum markers and imageological examination. Except CA19-9 and CEA, we can also combine LAPTM4B-35 with other serum biomarkers such as CD26, which has been explored extensively in PC patients by our team (22).
Conclusions
In summary, we evaluated the expression of LAPTM4B-35 in PC tissues and paired non-cancerous tissues and assessed the relationship of LAPTM4B-35 expression with clinicopathological features and post-operation survival, respectively. We found that LAPTM4B-35 might be a potential biomarker for PC at its early stage, and LAPTM4B-35 expression was correlated with favorable outcome of PC patients. Furthermore, LAPTM4B polymorphism has been found to be associated with multiple kinds of tumors and increasing susceptibility to tumors (23-27). So, our team is also examining LAPTM4B polymorphism of PC patients. We hope to combine the results of this study with LAPTM4B polymorphism and to find a junction point so as to better understand the mechanisms underlying development and progression of PC.
Acknowledgments
Funding: This work was supported by Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (approval No. XMLX201708); the Capital Health Research and Development Special (approval No. 2016-2-2151); the National Natural Science Funding (approval No. 81673500, 61372028, 61571437) and Capital Characteristic Clinical Application Research (approval No. Z161100000516065 and Z121107001012083) and International Science & Technology Cooperation Program of China (approval #: 2013DFG32720).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.07.11). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethical Committee of Peking University Cancer Hospital (No. 2016KT41) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Allison DC, Piantadosi S, Hruban RH, et al. DNA content and other factors associated with ten-year survival after resection of pancreatic carcinoma. J Surg Oncol 1998;67:151-9. [Crossref] [PubMed]
- Shao GZ, Zhou RL, Zhang QY, et al. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene 2003;22:5060-9. [Crossref] [PubMed]
- Liu X, Zhou R, Zhang Q, et al. Identification and characterization of LAPTM4B encoded by a human hepatocellular carcinoma-associated novel gene. Beijing Da Xue Xue Bao 2003;35:340-7. [PubMed]
- Yang H, Xiong F, Qi R, et al. LAPTM4B-35 is a novel prognostic factor of hepatocellular carcinoma. J Surg Oncol 2010;101:363-9. [Crossref] [PubMed]
- Yang H, Xiong F, Wei X, et al. Overexpression of LAPTM4B-35 promotes growth and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer Lett 2010;294:236-44. [Crossref] [PubMed]
- Yang H, Xiong FX, Lin M, et al. LAPTM4B-35 overexpression is a risk factor for tumor recurrence and poor prognosis in hepatocellular carcinoma. J Cancer Res Clin Oncol 2010;136:275-81. [Crossref] [PubMed]
- Xiao M, Jia S, Wang H, et al. Overexpression of LAPTM4B: an independent prognostic marker in breast cancer. J Cancer Res Clin Oncol 2013;139:661-7. [Crossref] [PubMed]
- Yin M, Lou C, Zhang W, et al. LAPTM4B overexpression is a novel independent prognostic marker for metastatic ovarian tumors. Int J Gynecol Cancer 2012;22:54-62. [Crossref] [PubMed]
- Cheng X, Zheng Z, Bu Z, et al. LAPTM4B-35, a cancer-related gene, is associated with poor prognosis in TNM stages I-III gastric cancer patients. PLoS One 2015;10:e0121559 [Crossref] [PubMed]
- Liu L, Xu X, Jing L, et al. Lysosomal-associated protein transmembrane 4 Beta-35 overexpression is a novel independent prognostic marker for gastric carcinoma. PLoS One 2015;10:e0118026 [Crossref] [PubMed]
- Meng F, Tan S, Liu T, et al. Predictive significance of combined LAPTM4B and VEGF expression in patients with cervical cancer. Tumour Biol 2016;37:4849-55. [Crossref] [PubMed]
- Kang Y, Yin M, Jiang W, et al. Overexpression of LAPTM4B-35 is associated with poor prognosis in colorectal carcinoma. Am J Surg 2012;204:677-83. [Crossref] [PubMed]
- Marullo M, Zuccato C, Mariotti C, et al. Expressed Alu repeats as a novel, reliable tool for normalization of real-time quantitative RT-PCR data. Genome Biol 2010;11:R9. [Crossref] [PubMed]
- Tan AC, Fan JB, Karikari C, et al. Allele-specific expression in the germline of patients with familial pancreatic cancer: an unbiased approach to cancer gene discovery. Cancer Biol Ther 2008;7:135-44. [Crossref] [PubMed]
- Liu X, Xiong F, Wei X, et al. LAPTM4B-35, a novel tetratransmembrane protein and its PPRP motif play critical roles in proliferation and metastatic potential of hepatocellular carcinoma cells. Cancer Sci 2009;100:2335-40. [Crossref] [PubMed]
- Maki Y, Fujimoto J, Lang W, et al. LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells. Sci Rep 2015;5:13846. [Crossref] [PubMed]
- Yang H, Lin M, Xiong F, et al. Combined lysosomal protein transmembrane 4 beta-35 and argininosuccinate synthetase expression predicts clinical outcome in hepatocellular carcinoma patients. Surg Today 2011;41:810-7. [Crossref] [PubMed]
- Li L, Wei XH, Pan YP, et al. LAPTM4B: a novel cancer-associated gene motivates multidrug resistance through efflux and activating PI3K/AKT signaling. Oncogene 2010;29:5785-95. [Crossref] [PubMed]
- Zhang G, Liang Y, Huang Y, et al. Elevated lysosome-associated protein transmembrane-4beta-35 is an independent prognostic marker in pancreatic carcinoma. J Int Med Res 2012;40:1275-83. [Crossref] [PubMed]
- Kasper G, Vogel A, Klaman I, et al. The human LAPTM4b transcript is upregulated in various types of solid tumours and seems to play a dual functional role during tumour progression. Cancer Lett 2005;224:93-103. [Crossref] [PubMed]
- Ye C, Tian X, Yan L, et al. Elevated serum CD26 level is associated with metastasis and post-operation survival in pancreatic cancer patients. Transl Cancer Res 2016;5:512-9. [Crossref]
- Cheng X, Tian X, Wu X, et al. Relationship between LAPTM4B Gene Polymorphism and Prognosis of Patients following Tumor Resection for Colorectal and Esophageal Cancers. PLoS One 2016;11:e0158715 [Crossref] [PubMed]
- Fan M, Liu Y, Zhou R, et al. Association of LAPTM4B gene polymorphism with breast cancer susceptibility. Cancer Epidemiol 2012;36:364-8. [Crossref] [PubMed]
- Tang H, Tian H, Yue W, et al. LAPTM4B polymorphism is associated with nonsmall cell lung cancer susceptibility and prognosis. Oncol Rep 2014;31:2454-60. [Crossref] [PubMed]
- Wang S, Zhang QY, Zhou RL. Relationship between LAPTM4B gene polymorphism and susceptibility of primary liver cancer. Ann Oncol 2012;23:1864-9. [Crossref] [PubMed]
- Zhang M, Zhou R, Xu J, et al. Relationship Between LAPTM4B Gene Polymorphism and Susceptibility of Malignant Melanoma in Chinese Patients. Transl Oncol 2014;7:638-43. [Crossref] [PubMed]


