
siRNA-mediated silencing of FOXQ1 expression inhibits cell proliferation and induces apoptosis in human osteosarcoma cells
Introduction
Osteosarcoma (OS) is known as the most common malignant primary bone tumor in children and young adults (1,2). It causes death in patients as the main results of the high degree of malignancy and early occurrence of metastasis, with the 5-year survival rate of the non-metastatic patients reaching 65% (3). Therefore, it is in urgent need to identify novel diagnostic and prognostic biomarkers and potential therapeutic targets to improve the prognosis in patients with OS. Although numerous genetic alterations have been reported to be associated with OS, molecular mechanisms underlying OS pathogenesis remain to be clarified. Forkhead box Q1 (FOXQ1) is a member of the forkhead transcription factor family, which is 2,338 base pairs in length and located in chromosome 6p25.3 (4). FOXQ1 is involved in chemoresistance, epithelial-mesenchymal transition, cell cycle progression, apoptosis, invasion, and metastasis of multiple cancers (5-8). Recently, studies have shown that FOXQ1 is overexpressed in lung cancer, breast cancer, and prostate cancer (9,10). In this study, we investigated the effect of FOXQ1 on cell proliferation and apoptosis in OS cells by RNA interference.
Methods
Human OS cell lines
U2OS cells were preserved by the Department of Orthopaedic Surgery, Tangdu Hospital, the Fourth Military Medical University. U2OS cells were cultured in MEM medium (Corning, USA) contained 10% fetal bovine serum (Gibco, USA) at 37 °C in humidified 5% CO2.
siRNA transfection
Specific siRNA directed against FOXQ1 gene was designed using the sense 5'-GGACACCUCCCTACAGGUCTT-3' and antisense 5'-ACCGCCACGTGAAGGGTAATT-3'. The negative control siRNA (NC-siRNA) was designed using the sense 5'-ATCCCCGAACGUGACACGUAT-3'. Before transfection, U2OS cells were plated in six-well plates at a density of 1×105 cells per well. When cell density is up to 60%, transfection assay was performed with FOXQ1-siRNA or NC-siRNA (the final concentration of siRNA was 100 nM) using Lipofectamine TM2000 Reagent (Invitrogen, USA) according to the manufacturer’s instructions. After incubation for 8 h, cells were rinsed and replenished with MEM containing 10% FBS. After transfection for 48 h or the indicated time points, cells were collected for further experiments. The experiment was replicated three times.
Real-time qRT-PCR
Cells were collected 48 h after transfection and total RNA was isolation. Real-time qRT-PCR assay was performed using SYBR Green dye. The qPCR reaction amplification conditions were 94 °C for 8 min, 95 °C for 10 s, 60 °C for 1 min for 35 cycles, and 72 °C for 45 s. FOXQ1 mRNA expression was measured using the sense 5'-G-AAGAACTCCATCCGCCACA-3' and antisense 5'-GCCT-TAAACACCTGGTCCAATGTC-3' primers. The β-actin was used as a control using sense 5'-TGGTTACCCATTGGGAACTGTC-3' and antisense 5'-TCACGGCCTGTTGTGTAGGTC-3' primers. Relative expression was calculated using the 2-∆∆Ct method and normalized to the expression of β-actin.
Western blotting
U2OS cells were harvested after transfection for 48 h. Cellular protein isolation and protein concentration determination were performed for Western blotting. Proteins were electrophoresed on 12% SDS-PAGE and transferred into nitrocellulose membranes at 100 V for 90 min. Membranes were blocked with 5% nonfat milk in TBST at room temperature for 2 h and incubated respectively with rabbit anti-FOXQ1 antibody and rabbit anti-β-actin antibody (CST, USA) at 4 °C overnight with gentle rocking. After washing with TBST, the membranes were incubated with the appropriate HRP-conjugated secondary antibodies [goat anti-rabbit IgG (Pierce, USA)] at room temperature for 2 h. After extensive washing with TBST, proteins were visualized by ECL detection. The gray values were analyzed using Quantity One software, and FOXQ1 expression was represented by the gray value ratio of FOXQ1 to β-actin.
Cell proliferation assay
The three groups of cells were harvested and seeded at a density of 0.8×104 cells per well in 96-well plates and cultured at 37 °C overnight. Each group was made in quadruplicating. Transfection was performed with FOXQ1-siRNA and NC-siRNA separately as described above. After 24, 48, 72, and 96 h treatment, 20 µL of MTT reagent was added into each well and the cells were incubated at 37 °C for 4 h. Then 200 µL of dimethyl sulfoxide (DMSO) was added into each well, and the absorbance values were measured at 490 nm in an automatic microplate reader. The experiment was replicated three times.
Apoptosis detection
Three groups of cells were harvested 72 h after transfection. Double staining with annexin V and PI was conducted according to the manufacturer’s protocol. The stained cells were detected using FACS. The experiment was replicated three times.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 software (SPSS, Chicago, IL, USA). The results were presented as the mean ± standard error. Quantitative data were performed by one-way ANOVA. Statistical multiple comparisons were performed with LSD test. P<0.05 was considered statistically significant.
Results
Special siRNA represses the expression of FOXQ1 mRNA expression
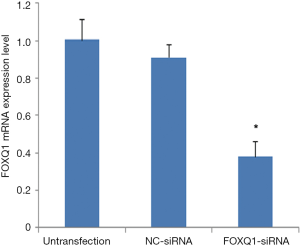
To explore the role of FOXQ1 in OS, inhibition of FOXQ1 by RNA inference is established in U2OS cells, as a high expression of FOXQ1 was detected in this cell line. We first performed the real-time qRT-PCR assay to determine the knockdown efficacy. The result showed that FOXQ1 mRNA expression was significantly lower in FOXQ1-siRNA group compared with those in the other two groups (P<0.05, Figure 1). This result indicated that siRNA transfection effectively silencing FOXQ1 expression.
siRNA-mediated silencing suppress FOXQ1 protein expression
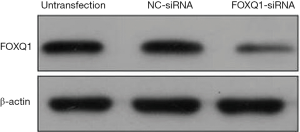
Furthermore, western blotting analysis showed that the protein level of FOXQ1 was remarkably depressed in FOXQ1-siRNA group compared with those in NC-siRNA group and untransfection group (Figure 2). This data confirmed that FOXQ1 expression was repressed effectively by siRNA-mediated silencing.
Downregulation of FOXQ1 expression inhibits cell proliferation
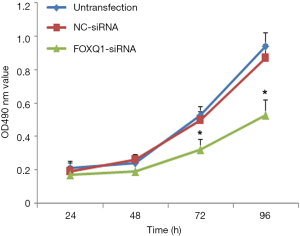
To assess the effect of FOXQ1 on the proliferation of U2OS cells, the cell viability of U2OS cells in untransfection group, NC-siRNA group, and FOXQ1-siRNA group were detected using MTT assay. The result showed that U2OS cells in FOXQ1-siRNA group proliferated more slowly than those in the other two groups (Figure 3). This result suggested that downregulation of FOXQ1 expression reduced U2OS cell proliferation in vitro.
Inhibition of FOXQ1 promotes apoptosis in U2OS cell line
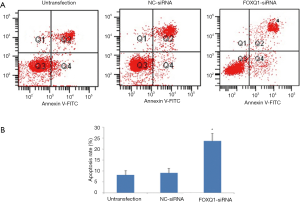
Apoptosis is critical for cell proliferation. We next investigated whether downregulation of FOX1 has an effect on U2OS cell proliferation. The result of flow cytometry analysis showed that the apoptosis rate of U2OS cells in FOXQ1-siRNA group was significantly increased compared to those in NC-siRNA group and untransfection group (Figure 4), suggesting that inhibition of FOXQ1 expression enhanced the apoptosis of U2OS cells.
Discussion
FOXQ1 is a member of the FOX gene family, which is involved in embryonic development, cell cycle regulation, tissue-specific gene expression, cell signaling, and tumorigenesis (11-13). Recently, studies showed that FOXQ1 overexpressed in tumors is important in the progression and malignant proliferation of cancer cells (14). It has been reported that high FOXQ1expression is correlated with large tumor diameter, high serum α-fetoprotein levels, and later stage grouping with tumor node metastasis classification (15). Feng et al. reported that FOXQ1 regulates EMT and increases chemosensitivity in non-small cell lung cancer cells (6), while Christensen et al. showed that the overexpression of FOXQ1 promotes cell proliferation, migration, and invasion of breast cancer cells (16). Pei et al. also demonstrated that FOXQ1 negatively modulates CDH1 to promote esophageal cancer proliferation and metastasis. These results suggest that FOXQ1 plays an important role in cancers.
Currently, despite the progress on OS treatment, the 5-year survival rate of Chinese patients remains to be 63.6%, and median survival rate to be 64.0% (17). Therefore, it is urgent to search novel molecular targets for the treatment of OS. A number of studies indicate that FOXQ1 is critical in tumorigenesis (18); however, the function and molecular mechanism of FOXQ1 involved in OS are still unclear. RNAi interference is a posttranscriptional sequence-selective technique for the silencing of genes by suppressing gene expression in a sequence-specific method (19). Zhang et al. have reported that knockdown of FOXQ1 by siRNA significantly suppresses laryngeal cancer cell growth (Zhang, Li et al., 2015). In this study, we found that FOXQ1 expression was detectable in U2OS cells and that siRNA-mediated silencing of FOXQ1 expression remarkably inhibited U2OS cell proliferation. Moreover, we found that knockdown of FOXQ1 induced apoptosis in U2OS cells, which was consistent with previous report that inhibition of FOXQ1 enhanced apoptosis of prostate cancer cells (Zhang, Wang et al., 2016). It has been reported that FOXM1, another member of the FOX gene family, blocks cell cycle and promotes apoptosis in company with PIK-1, Survivin and CDC25B (Xu et al., 2012). Whether FOXQ1 regulates OS cell apoptosis in a similar way is needed to be investigated in further studies.
In summary, our data provided evidence that siRNA-mediated targeting depression of FOXQ1 expression inhibits the proliferation and induces apoptosis in OS cells. Thus, FOXQ1 might be a novel molecular target for OS treatment.
Acknowledgments
The authors thank Xing Huang and Fei Ding for cell culture and technique support.
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.27). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang J, Wang X, Wu W, et al. Expression of the Nrf2 and Keap1 proteins and their clinical significance in osteosarcoma. Biochem Biophys Res Commun 2016;473:42-6. [Crossref] [PubMed]
- Sharma T, Hamilton R, Mandal CC. miR-214: a potential biomarker and therapeutic for different cancers. Future Oncol 2015;11:349-63. [Crossref] [PubMed]
- Peng X, Luo Z, Kang Q, et al. FOXQ1 mediates the crosstalk between TGF-beta and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther 2015;16:1099-109. [Crossref] [PubMed]
- Qin J, Xu Y, Li X, et al. Effects of lentiviral-mediated Foxp1 and Foxq1 RNAi on the hepatocarcinoma cell. Exp Mol Pathol 2014;96:1-8. [Crossref] [PubMed]
- Fan DM, Feng XS, Qi PW, et al. Forkhead factor FOXQ1 promotes TGF-beta1 expression and induces epithelial-mesenchymal transition. Mol Cell Biochem 2014;397:179-86. [Crossref] [PubMed]
- Feng J, Xu L, Ni S, et al. Involvement of FoxQ1 in NSCLC through regulating EMT and increasing chemosensitivity. Oncotarget 2014;5:9689-702. [Crossref] [PubMed]
- Meng F, Speyer CL, Zhang B, et al. PDGFRalpha and beta play critical roles in mediating Foxq1-driven breast cancer stemness and chemoresistance. Cancer Res 2015;75:584-93. [Crossref] [PubMed]
- Peng XH, Huang HR, Lu J, et al. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol Cancer 2014;13:186. [Crossref] [PubMed]
- Feng J, Zhang X, Zhu H, et al. FoxQ1 overexpression influences poor prognosis in non-small cell lung cancer, associates with the phenomenon of EMT. PLoS One 2012;7:e39937 [Crossref] [PubMed]
- Ross JB, Huh D, Noble LB, et al. Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nat Cell Biol 2015;17:651-64. [Crossref] [PubMed]
- Pei Y, Wang P, Liu H, et al. FOXQ1 promotes esophageal cancer proliferation and metastasis by negatively modulating CDH1. Biomed Pharmacother 2015;74:89-94. [Crossref] [PubMed]
- Tang B, Becanovic K, Desplats PA, et al. Forkhead box protein p1 is a transcriptional repressor of immune signaling in the CNS: implications for transcriptional dysregulation in Huntington disease. Hum Mol Genet 2012;21:3097-111. [Crossref] [PubMed]
- Betts L, Merenbloom BK, Lord ST. The structure of fibrinogen fragment D with the 'A' knob peptide GPRVVE. J Thromb Haemost 2006;4:1139-41. [Crossref] [PubMed]
- Zhan HX, Xu JW, Wang L, et al. FoxQ1 is a Novel Molecular Target for Pancreatic Cancer and is Associated with Poor Prognosis. Curr Mol Med 2015;15:469-77. [Crossref] [PubMed]
- Wang W, He S, Ji J, et al. The prognostic significance of FOXQ1 oncogene overexpression in human hepatocellular carcinoma. Pathol Res Pract 2013;209:353-8. [Crossref] [PubMed]
- Christensen J, Bentz S, Sengstag T, et al. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS One 2013;8:e60051 [Crossref] [PubMed]
- Li X, Ashana AO, Moretti VM, et al. The relation of tumour necrosis and survival in patients with osteosarcoma. Int Orthop 2011;35:1847-53. [Crossref] [PubMed]
- Bao B, Azmi AS, Aboukameel A, et al. Pancreatic cancer stem-like cells display aggressive behavior mediated via activation of FoxQ1. J Biol Chem 2014;289:14520-33. [Crossref] [PubMed]
- Qiao Y, Jiang X, Lee ST, et al. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res 2011;71:3076-86. [Crossref] [PubMed]


