
Prognostic value of NF-κB interacting long non-coding RNA expression in lung adenocarcinoma and lung squamous cell cancer: a study based on the cancer genome atlas datasets
IntroductionOther Section
Lung cancer is the most frequently diagnosed cancer (1). lung adenocarcinoma (LUAD) has become the most common type of lung cancer in both smokers [replacing lung squamous cell cancer (LUSC)] and lifelong nonsmokers (2). LUAD and LUSC originate from different cells and have several major differences, not only in their biological patterns but also in their molecular characteristics and, most importantly, the therapeutic strategies required to treat them (3-6). Predictive biomarkers include the ALK fusion oncogene (fusion between ALK and other genes) (7) and the sensitizing EGFR mutation (8), which are not routinely found in patients with squamous cell carcinoma (9). The NCCN strongly endorses broader molecular profiling to identify rare driver mutations to ensure that patients receive the most appropriate treatment (6). Therefore, comprehensive investigations into the differences of molecular characteristics and mechanisms of these two major subtypes of lung cancer are required.
The lncRNAs, defined as RNA molecules >200 nucleotides in length, have been shown to be involved in the regulation of gene expression in both tumor-suppressive and oncogenic pathways at the epigenetic level, transcriptional level, and posttranscriptional level (10-13). Major tumor suppressor and oncogenic lncRNAs in lung cancer include BANCR (BRAF-activated non-protein coding RNA), PANDA (P21-associated noncoding RNA DNA damage-activated) and tumor oncogenic lncRNA MALAT1, HOTAIR (lncRNA homeotic genes HOX transcript antisense RNA), CCAT2 (colon cancer associated transcript-2) (14). The most well-characterized lncRNA reported in lung cancer is metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which is associated with high metastatic potential and poor patient prognosis in non-small cell lung cancer (NSCLC) patients with and without metastatic tumors (15). NF-κB interacting long non-coding RNA (NKILA), a lncRNA encoded by a gene at chromosome 20q13, has been reported to indicate the development and metastasis of several cancers, such as breast cancer, malignant melanoma, tongue squamous cell carcinoma of the tongue (TSCC) and NSCLC (16-19). In NSCLC, Lu et al. proposed that the expression of NKILA was regulated through classical TGF-β signal pathway, which subsequently inhibited migration and invasion of NSCLC cells through interfering NF-κB/Snail signal pathway in NSCLC cells (17). However, the effects of NKILA in subgroups of NSCLC (LUAD and LUSC) was not entirely clear.
In 2006, the Cancer Genome Atlas (TCGA), a project initiated by the National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI), aimed to catalogue the mutations responsible for cancer, and their DNA and RNA sequencing data are publicly accessible to the community and researchers. The Atlas of ncRNA in Cancer (TANRIC, http://ibl.mdanderson.org/tanric/_design/basic/query.html) is an open-access resource for the interactive exploration of lncRNAs in cancer. This atlas characterizes the expression profiles of lncRNAs in large patient cohorts of 20 cancer types including TCGA, Cancer Cell Line Encyclopedia (CCLE) and other independent datasets, which uses RPKM (Reads Per Kilobase per Million mapped reads) to quantify the expression levels of lncRNAs based on BAM (Binary Alignment/Map) files. Both TCGA and TANRIC collect data on LUAD and LUSC respectively, enabling us to explore their differences using more samples.
Because of the differences of molecular characteristics between LUAD and LUSC mentioned above, we investigate if there is any difference in the expression levels of NKILA between them. We would like to analyze the diagnostic value of NKILA in LUAD and LUSC in order to further evaluate the prognostic value of NKILA expression in NSCLC based on these two open databases (TCGA and TANRIC).
MethodsOther Section
Data collection from the TCGA database
The expression of NKILA in lung cancers (including LUAD and LUSC) was extracted from TANRIC by the position (chr20:56285239-56287836; hg19). A total of 546 entries of LUAD and 237 entries of LUSC were analyzed. Among the entries, 58 LUAD and 17 LUSC samples were pairs of both tumor tissues and normal tissues. The clinical data of those samples were extracted from the TCGA project. A total of 476 LUAD and 220 LUSC subjects with both survival and NKILA expression data were included. Samples were excluded if there was no data of age or stages, and one sample each of American Indian or Alaska native patients were also excluded. A total of 450 LUAD and 211 LUSC samples were enrolled in the subsequent analyses. The NKILA expression levels were dichotomized into high expression and low expression groups by the median value, respectively. The survival curves were plotted and tested using the Kaplan-Meier method and log-rank test.
qRT-PCR
In this study, six formalin-fixed, paraffin-embedded (FFPE) LUAD tissues, including their adjacent healthy non-tumorous lung tissues, were collected from the Department of Pathology of Tongji Hospital at Tongji Medical College, Huazhong University of Science and Technology (Wuhan, Hubei, China), from January 2012 to February 2014. The research protocol for this study was approved by the Ethics Committee. The total RNA was extracted from LUAD tissue samples using RNAiso Plus (Takara, Japan) and later reverse-transcribed with a PrimeScript™ RT Master Mix (Takara, Japan) according to the manufacturer’s instructions. qRT-PCR was performed to quantify the expression levels of NKILA using the Biosystems 7300 Real-Time PCR System (Life Technologies) and the 2× PCR Master mix (Takara, Japan). The NKILA primer sequences used were as follows: forward, 5'-AACCAAACCTACCCACAACG-3'; reverse, 5'-ACCACTAAGTCAATCCCAGGTG-3'. β-actin was evaluated as a housekeeping gene. The β-actin primers used were forward, 5'-GCAAGCAGGACTATGACGAG-3'; reverse, 5'-CAAATAAAGCCATGCCAATC-3'. The total reaction volume was 10 µL, including 5 µL of SYBR-Green qPCR Mix (Takara), 1 µL of cDNA, 0.2 µL of ROX reference dye, 0.4 µL of forward and reverse primers at 0.4 µmol/L, respectively, and 3 µL of diethyl pyrocarbonate (DEPC) water. The reaction conditions were as follows: 50 °C for 2 min for 1 cycle (first stage); 95 °C for 2 min for 1 cycle (second stage); 95 °C for 15 s; 60 °C for 30 s for 40 cycles (third stage). Melting curve analysis involved one cycle of 95 °C for 15 s, 60 °C for l min, 95 °C for 15 s, and 60 °C for 15 s. Every reaction mixture was completed in triplicate and qRT-PCR was also tripled for each sample and all cell groups. The expression levels of NKILA were normalized to the internal control β-actin reference to obtain the relative threshold cycle (ΔCt). The relative levels were calculated by the comparative Ct (ΔΔCt) method, and the relative expression fold (2−ΔΔCt) was calculated.
Statistical analysis
Statistical analysis was carried out by SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA). All data are displayed as the mean ± SEM from each group. Two-tailed Student’s t-test was utilized to analyze the significance of the difference between 2 groups. One-way ANOVA was performed to test the significance of the difference among 3 or more groups. To differentiate the expression data between the controls and tumor tissues, the diagnostic value was identified using a receiver operator characteristic (ROC) curve. Kaplan-Meier survival curves were implemented for the relationship between the NKILA expression and the overall survival (OS)/5-year survival in cancer patients. OS was measured from the beginning of the study to the death of the patient (from any cause) or to the last follow-up time for the surviving patients. A two-sided P<0.05 was considered to reveal a statistically significant difference.
ResultsOther Section
Patient characteristics of the selected data from TCGA
In total, 696 samples were obtained from the TCGA database. After we excluded those cases lacking expression values or complete clinical information, 661 samples were included in the subsequent analyses. Clinical information for all 661 samples, including 450 LUAD patients and 211 LUSC patients, was reviewed. In LUAD, there were 202 males (44.9%) and 248 females (55.1%) aged from 38 to 88 years (mean: 67 years), The patients providing the LUAD samples were mainly white (91.4%). Of the 450 cases of LUAD, 245 (54.4%) patients were diagnosed with stage I LUAD, and the number of stage II, III, IV patients were 105 (23.3%), 77 (17.1%) and 23 (5.1%), respectively. It was noteworthy that the majority of these patients were diagnosed at relatively early stages. There were relatively more male LUSC patients (153, 72.5%) than LUAD patients (202, 44.9%). Other characteristics of LUSC were similar to those of LUAD. Smokers constituted the majority of NSCLC patients, both in LUAD and LUSC. Detailed information of both the LUAD and LUSC patients are summarized in Table 1.
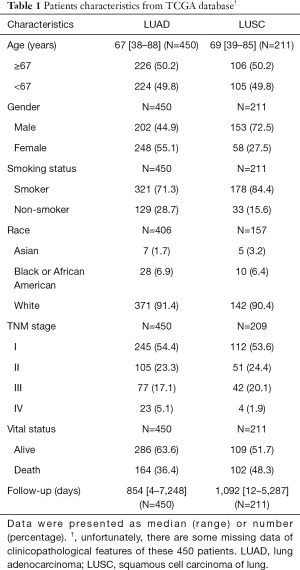
Full table
NKILA was upregulated in LUAD and LUSC cancer tissues
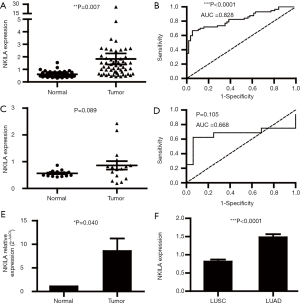
Among the LUAD and LUSC patients, we observed 57 LUAD and 17 LUSC patients with both data of cancer and normal tissues. First, we evaluated the expression of NKILA in 57 pairs of LUAD cancer tissues and their adjacent normal tissues. The expression of NKILA was significantly upregulated in LUAD cancer tissues compared with the normal tissues (1.833±0.440 vs. 0.612±0.028; P=0.007; Figure 1A). In Figure 1B, the ROC curve was utilized to determine the diagnostic value in LUAD. The area under the curve (AUC) of NKILA was 0.828 (95% CI: 0.751–0.905; P<0.0001), with a sensitivity and specificity of 66.67% and 94.74%, respectively. Unfortunately, although a similar trend could be detected in LUSC, there was no significant difference between the cancer and normal tissues (0.863±0.158 vs. 0.563±0.028; P=0.089) (Figure 1C). In addition, the ROC curve analysis showed that the AUC was 0.668 (95% CI: 0.454–0.882; P=0.105; Figure 1D). Small samples of LUAD (n=6) collected from the Tongji Hospital were used to verify the high expression levels of NKILA by RT-qPCR, and the relative NKILA expression in cancer tissues compared with normal tissues calculated by the 2−ΔΔCt method was 8.495±2.719 vs. 1.002±0.001 (P=0.040; Figure 1E). According to the results above, NKILA was upregulated in LUAD and can be used as a diagnostic marker for LUAD, rather than LUSC. Finally, we compared the expression of NKILA between LUAD and LUSC, and we observed that it was significantly higher in LUAD (1.482±0.083 vs. 0.815±0.055, P<0.0001; Figure 1F), which further suggested that LUAD and LUSC were different at the molecular level and should thus be analyzed separately.
NKILA expression and clinical characteristics in LUAD and LUSC
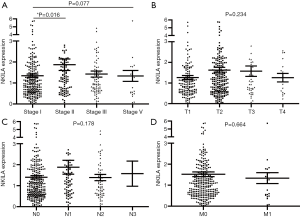
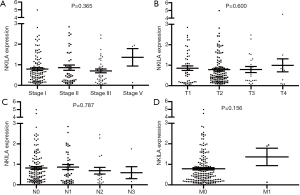
The statistical analyses revealed no correlation between the expression level of NKILA and age, gender, smoking status, race, TNM stage, and the presences of either lymph node metastasis or distant metastasis (Tables 2,3). The correlation of NKILA expression and TNM stages is shown in Figures 2,3. There was no statistic difference between the T, N, M stages of LUSC, but we found significant differences between stage I and stage II in LUAD (1.347±0.080 vs. 1.870±0.273, P=0.016; Figure 2A).
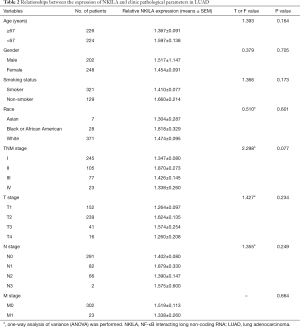
Full table
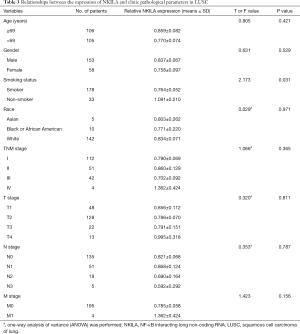
Full table
High NKILA expression was correlated with poor survival in LUAD
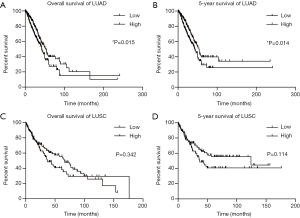
To assess the prognostic value of NKILA expression in LUAD and LUSC, we next analyzed its correlation with the OS and 5-year survival. In LUAD patients, the expression of NKILA was categorized as either low (n=225) or high (n=225) expression levels the median expression as a cutoff (Figure 4A,B). Higher levels of NKILA expression were associated with a reduced OS, with median survival of 42.17 months (95% CI: 36.27–48.03 months), compared to the period of 53.33 months (95% CI: 43.70–62.93 months) for the low NKILA expression group (log-rank, P=0.015; Figure 4A). The 5-year survival of the NKILA high expression group was also lower than the low expression group (log-rank, P=0.014; Figure 4B). In LUSC, the OS and 5-year survival revealed the same trend but there was no significant difference (OS: P=0.342, Figure 4C; 5-year survival: P=0.114, Figure 4D). The median survival rates of the high and low expression groups in LUSC were 36.90 months (95% CI: 24.57–49.20 months) and 65.83 months (95% CI: 47.73–83.90 months), respectively. Thus, high NKILA expression was correlated with poor survival in LUAD, making it a reasonable potential prognostic marker for LUAD.
DiscussionOther Section
Lung cancer is one of the most-deadly diseases worldwide with a 5-year survival rate at approximately 15% being observed (1). LUAD is the single most common type of lung cancer, accounting for approximately 40% of all lung cancers (20). Significant efforts have been made to identify the molecular changes that could be used for early diagnosis and to predict the prognosis of cancers and guide clinical treatment (21-25). In the past years, lncRNAs have gradually been elucidated as a key component of cancers. MALAT1 is a candidate of biomarker in liquid biopsy for the diagnosis of NSCLCs. High MALAT1 expression in formalin-fixed paraffin-embedded specimens indicates a higher mortality in NSCLCs. Liu and colleagues reported that elevated HOTAIR expression was associated with cisplatin resistance in NSCLC, and showed that HOTAIR expression was directly related to Kruppel-like factor 4 (KLF4) expression, suggesting a new therapeutic target for drug-resistance patients with NSCLC (26). Taken together, the results of these studies indicate that lncRNAs can be useful biomarker for lung cancer.
In this study, we tested the expression of NKILA in LUAD and LUSC samples and their surrounding normal lung tissues. The results showed an upregulation of NKILA in tumor tissues of both LUAD and LUSC patients, but this difference was statistically significant only in LUAD patients. The mean expression of NKILA in LUAD tumor tissues was approximately 3 times higher than that in normal tissues. Thus, we think that NKILA may be a potential biomarker for LUAD. Contrary to our results, Lu and colleagues reported that NKILA was down-regulated in NSCLC cancer tissues compared with matched adjacent noncancerous tissues, and lower NKILA expression in the tumor tissues was significantly correlated with lymph node metastasis and advanced TNM stages. This process was primarily regulated by TGF-β (17). The different racial background of patients may explain these seemingly conflicting results. Lu’s study focused on Asians, but our data from TCGA included mostly white individuals. Second, Lu et al. mixed the LUAD and LUSC samples, while we analyzed them separately thus observed that NKILA was higher in LUAD than LUSC, which may again widen the discrepancy. Furthermore, the majority of samples were in stage I or II without lymph node or distant metastasis, but most patients diagnosed in China were already in the advanced stages (27). We thus speculated that NKILA may be highly correlated with the early stages of NSCLC, which also needs further investigation. Of course, the number of LUSC patients was only 17, and more samples are thus needed to support our conclusion.
Second, we observed that mean expression of NKILA in LUAD was approximately 2 times higher than that in LUSC. Previous studies have found the distinctive expression pattern in LUAD and LUSC. Wei et al. identified a set of 27 lncRNAs which was just dysregulated in LUAD, as well as a panel of 68 lncRNAs only dysregulated in LUSC compared with normal lung tissues (28). The rs3200401 T allele located on the lncRNA MALAT1 was associated with a better survival for advanced LUAD patients, but not with LUSC patients (29). LncRNAs are stable even in body fluids and show tissue-specific expression. In this background, NKILA can be a candidate marker for screening and early diagnosis of lung cancer. The distinct NKILA expression in LUAD and LUSC will help us understand the molecular difference between the two subsets which will, in turn, further guide improved clinical treatment.
Most importantly, we first reported in LUAD that the OS of patients with lower NKILA expression levels was significantly higher than that of patients with higher NKILA expression levels. These findings suggest that NKILA can be a useful biomarker for the prognosis of LUAD patients. Liu and colleagues reported that NKILA could be upregulated by NF-κB and bound to NF-κB/IκB, serving to directly mask phosphorylation motifs of IκB and thereby inhibiting IKK-induced IκB phosphorylation and NF-κB activation in normal breast tissues (19). But in breast cancer, NKILA was significantly reduced by miR-103/107-mediated degradation, which resulted in abnormal NF-κB activation and thus contribute to cancer metastasis and poor patient outcome. Huang et al. (18) and Bian et al. (16) reported that low NKILA expression was significantly correlated with tumor metastasis and poor prognosis in TSCC and malignant melanoma patients, respectively. Even though convincing experimental data have identified NF-κB as a critical promoter of cancer development, data have also shown that NF-κB activity enhances tumor cell sensitivity to apoptosis and senescence (30-32). Liu et al. proposed that canonical NF-κB is a Fas transcription activator and that alternative NF-κB is a Fas transcription repressor (33). If NF-κB promotes Fas-mediated apoptosis in cancer cells, the inhibition of NF-κB may suppress Fas-mediated apoptosis to impair host immune cell-mediated tumor suppression (34,35). Li et al. recently reported in NSCLC that canonical NF-κB signaling was continuously attenuated in the cancer group during peripheral blood monocyte (MoDCs) differentiation, likely resulting in the transcriptional suppression of CIITA, a crucial trans-activator of MHC class II genes, leading to the further impairment of antigen presentation (36). This finding means that the inhibition of NF-κB by NKILA does not definitely lead to cancer suppression. NKILA may also function as a pro-tumoral factor through NF-κB/Fas-mediated apoptosis or other signaling pathways besides NF-κB. Of course, further studies are needed to reveal to the role of NKILA and NF-κB in NSCLC.
It was reported that NKILA is the antisense of PMEPA1 (prostate transmembrane protein, androgen induced 1) (37), a highly conserved gene through evolution and one of the most highly inducible genes in invasive cancers. The expression of PMEPA1 is associated with several tumors and it encodes a protein that participated in immune pathways (38-40). Vo Nguyen et al. proposed that highly expressed PMEPA1 suppressed levels of Smad phosphorylation in lung cancer cells and reduced the growth inhibitory effects of TGF-β/Smad signaling (41). There possibly are interactive effects between PMEPA1 and NKILA in tumor progression that have not been elucidated yet.
Our study has some limitations. First, no likely mechanism for NKILA has been delineated. Second, although TCGA is one of the most commonly used database in human cancer profiling, it does not represent all of the lung cancer patients, especially from an Asian population. In China, although new techniques have been developed, the majority of patients were diagnosed at a relatively advanced stage, which differed greatly from our samples. The samples size to verify the result of TCGA database was small. Third, NKILA may play different roles on each stage of the tumor, which was not taken into consideration in our study. Thus, further large-scale studies including Asians are needed to confirm our findings.
In conclusion, by comparing lncRNA expression profiles in cancerous tissues with those of normal lung tissues based on validation from TCGA datasets, we observed that aberrantly expressed NKILA is involved in LUAD and LUSC. Additionally, we identified NKILA as a prognostic marker for LUAD.
AcknowledgmentsOther Section
The authors are grateful to Dr. Weining Xiong, Dr. Yong Cao and Dr. Dingsheng Jiang for their helpful instructions and for approving the final approval of the version to be published.
Funding: None.
FootnoteOther Section
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethical Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (ID: TJ-C20121219). Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Jemal A, Ward EM, Johnson CJ, et al. Annual Report to the Nation on the Status of Cancer, 1975-2014, Featuring Survival. J Natl Cancer Inst 2017;109. [PubMed]
- Cheng TY, Cramb SM, Baade PD, et al. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J Thorac Oncol 2016;11:1653-71. [Crossref] [PubMed]
- Faruki H, Mayhew GM, Serody JS, et al. Lung Adenocarcinoma and Squamous Cell Carcinoma Gene Expression Subtypes Demonstrate Significant Differences in Tumor Immune Landscape. J Thorac Oncol 2017;12:943-53. [Crossref] [PubMed]
- Daraselia N, Wang Y, Budoff A, et al. Molecular signature and pathway analysis of human primary squamous and adenocarcinoma lung cancers. Am J Cancer Res 2012;2:93-103. [PubMed]
- Patnaik S, Mallick R, Kannisto E, et al. MiR-205 and MiR-375 microRNA assays to distinguish squamous cell carcinoma from adenocarcinoma in lung cancer biopsies. J Thorac Oncol 2015;10:446-53. [Crossref] [PubMed]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology. Non-Small Cell Lung Cancer, Version 8. 2017. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 2010;17:889-97. [Crossref] [PubMed]
- Warth A, Penzel R, Lindenmaier H, et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: patient outcome, interplay with morphology and immunophenotype. Eur Respir J 2014;43:872-83. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013;8:823-59. [Crossref] [PubMed]
- Li Y, Wang X. Role of long noncoding RNAs in malignant disease Mol Med Rep 2016;13:1463-9. (Review). [Crossref] [PubMed]
- Yu L, Fang F, Lu S, et al. lncRNA-HIT promotes cell proliferation of non-small cell lung cancer by association with E2F1. Cancer Gene Ther 2017;24:221-6. [Crossref] [PubMed]
- Xu MD, Wang Y, Weng W, et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin Cancer Res 2017;23:2071-80. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Inamura K. Major Tumor Suppressor and Oncogenic Non-Coding RNAs: Clinical Relevance in Lung Cancer. Cells 2017;6:E12 [Crossref] [PubMed]
- Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open 2015;5:e008653 [Crossref] [PubMed]
- Bian D, Gao C, Bao K, et al. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-kB. Am J Cancer Res 2017;7:28-40. [PubMed]
- Lu Z, Li Y, Wang J, et al. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-kappaB/Snail pathway. J Exp Clin Cancer Res 2017;36:54. [Crossref] [PubMed]
- Huang W, Cui X, Chen J, et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget 2016;7:62520-32. [PubMed]
- Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370-81. [Crossref] [PubMed]
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Wei S, Zhang ZY, Fu SL, et al. Hsa-miR-623 suppresses tumor progression in human lung adenocarcinoma. Cell Death Dis 2016;7:e2388 [Crossref] [PubMed]
- Nadal E, Zhong J, Lin J, et al. A MicroRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin Cancer Res 2014;20:3107-17. [Crossref] [PubMed]
- Rothschild SI, Tschan MP, Federzoni EA, et al. MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma. Oncogene 2012;31:4221-32. [Crossref] [PubMed]
- Kosari F, Ida CM, Aubry MC, et al. ASCL1 and RET expression defines a clinically relevant subgroup of lung adenocarcinoma characterized by neuroendocrine differentiation. Oncogene 2014;33:3776-83. [Crossref] [PubMed]
- Wang ZH, Li Z, Hu M, et al. Ovol2 gene inhibits the Epithelial-to-Mesenchymal Transition in lung adenocarcinoma by transcriptionally repressing Twist1. Gene 2017;600:1-8. [Crossref] [PubMed]
- Liu Z, Sun M, Lu K, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One 2013;8:e77293 [Crossref] [PubMed]
- Zheng R, Zeng H, Zuo T, et al. Lung cancer incidence and mortality in China, 2011. Thorac Cancer 2016;7:94-9. [Crossref] [PubMed]
- Wei Y, Zhang X. Transcriptome analysis of distinct long non-coding RNA transcriptional fingerprints in lung adenocarcinoma and squamous cell carcinoma. Tumour Biol 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Wang JZ, Xiang JJ, Wu LG, et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer 2017;17:167. [Crossref] [PubMed]
- Park MH, Hong JT. Roles of NF-κB in Cancer and Inflammatory Diseases and Their Therapeutic Approaches. Cells 2016;5:E15 [Crossref] [PubMed]
- Rinkenbaugh AL, Baldwin AS. The NF-κB Pathway and Cancer Stem Cells. Cells 2016;5:E16 [Crossref] [PubMed]
- Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest 2001;107:241-6. [Crossref] [PubMed]
- Liu F, Bardhan K, Yang D, et al. NF-κB directly regulates Fas transcription to modulate Fas-mediated apoptosis and tumor suppression. J Biol Chem 2012;287:25530-40. [Crossref] [PubMed]
- Varfolomeev E, Blankenship JW, Wayson SM, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 2007;131:669-81. [Crossref] [PubMed]
- Chien Y, Scuoppo C, Wang X, et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev 2011;25:2125-36. [Crossref] [PubMed]
- Li R, Fang F, Jiang M, et al. STAT3 and NF-κB are Simultaneously Suppressed in Dendritic Cells in Lung Cancer. Sci Rep 2017;7:45395. [Crossref] [PubMed]
- Dijkstra JM, Alexander DB. The " NF-k B interacting long noncoding RNA" (NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Res 2015;4:96. [PubMed]
- Fournier PG, Juarez P, Jiang G, et al. The TGF-beta Signaling Regulator PMEPA1 Suppresses Prostate Cancer Metastases to Bone. Cancer Cell 2015;27:809-21. [Crossref] [PubMed]
- Singha PK, Pandeswara S, Geng H, et al. TGF-beta induced TMEPAI/PMEPA1 inhibits canonical Smad signaling through R-Smad sequestration and promotes non-canonical PI3K/Akt signaling by reducing PTEN in triple negative breast cancer. Genes Cancer 2014;5:320-36. [PubMed]
- Liu R, Zhou Z, Huang J, et al. PMEPA1 promotes androgen receptor-negative prostate cell proliferation through suppressing the Smad3/4-c-Myc-p21 Cip1 signaling pathway. J Pathol 2011;223:683-94. [Crossref] [PubMed]
- Vo Nguyen TT, Watanabe Y, Shiba A, et al. TMEPAI/PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci 2014;105:334-41. [Crossref] [PubMed]


