Construction of dysregulated long non-coding RNA-associated competing endogenous RNA network in uterine corpus endometrial carcinoma
Introduction
Uterine corpus endometrial carcinoma (UCEC), a considerable threat to the health of women all over the world, is stemming from the inner lining cells of the uterus (1,2). The prognosis of UCEC is poor especially in patients who happened recurrence or metastasis after radiotherapy or surgery. Previous studies have attempted to identify tumor biomarkers and have identified multiple UCEC biomarkers, such as activated leukocyte cell adhesion molecule (CAM) (3), L1 CAM, etc. (4). Up to now, there is no way to fully elucidate the pathogenesis of UCEC. Accordingly, identification of personalized treatment strategies is urgently needed, such as potential biomarkers and therapeutic targets.
In 2011, Salmena et al. proposed the hypothesis of competing endogenous RNA (ceRNA), which described a complicated post-transcriptional regulatory network. In this network, lncRNAs, mRNAs, and other RNAs can act as natural miRNA sponges, sharing one or more miRNA response elements (MREs) to restrain miRNA function (5). In the last few years, an increasing number of researches have confirmed that the lncRNA-miRNAs-mRNAs regulation network plays a crucial role in pathogenesis and progression of malignant tumors such as breast cancer, colorectal cancer, gastric cancer, liver cancer, lung cancer, oral cancer, and thyroid cancer (6-14).
Non-coding RNA molecules (ncRNAs) longer than 200 nucleotides (nt) were usually named as long non-coding RNAs (lncRNAs) which distinguished from short-chain ncRNAs (15). LncRNA as ceRNA can compete with miRNAs to regulate the level of the gene encoding protein and participate in regulating the cell biology. MiRNAs have become an indispensable part of the ceRNA network by competition with target mRNA, inhibition of mRNA expression (16). However, a complete genome-wide and large number of samples analysis of regulatory network relating to lncRNA-miRNA-mRNA of UCEC it is still lacking. The current study clarified how the UCEC-related lncRNAs serve as ceRNAs to regulate target genes and is involved in pathogenesis and prognosis of UCEC.
The Cancer Genome Atlas (TCGA) is a public dataset that includes widespread and coordinated genome alterations in as many as 33 different types of cancer, which has been applied to collect, select, and analyze genomic changes in all kinds of cancers. As far as we know, ceRNA network of UCEC patients has not been constructed up to now. Therefore, the purpose of this current research was to determine a multi-RNA-based classifier based on the study of the lncRNA, miRNA and mRNA profiles of UCEC patients.
Methods
Data acquisition and selection
RNA expression profiles with clinical information and data were entirely downloaded from TCGA database. For good measure, the level 3 mRNASeq gene expression profiles, miRNAseq data of samples, and clinical data of those patients were downloaded by the Data Transfer Tool (provided by GDC Apps) (https://tcga-data.nci.nih.gov/). The sequencing platforms of the sequenced data were Illumina HiSeq RNASeq and Illumina HiSeq_miRNA-Seq platform. The downloaded data consists of many individual data files, each representing a tissue sample. A total of 552 UCEC samples (cohort T) and 35 normal control samples (cohort N) were included in the dataset. Our research conforms the publication guidelines offered by TCGA (http://cancergenome.nih.gov/publications/ publication guidelines). TCGA dataset collected and processed patients’ genome data under the precondition of abiding by the data access policies which have been authorized by its ethics committee, as a result, our current research did not need further ethical approval.
Differentially expressed analysis
Differential expressed lncRNAs, mRNAs and miRNAs (DElncRNAs, DEmRNAs, DEmiRNAs) between cohort T and cohort N were identified by differentially expression analysis through the R/Bioconductor package of edgeR (R version 3.4.4) (17). The cut-off values were |log2FC| >2 and FDR <0.01 (FC, fold change; FDR, false discovery rate).
Functional enrichment analysis
The function and pathway enrichment analyses were conducted by Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources (https://david.ncifcrf.gov/). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were researched. The criteria were set at P<0.05.
Constructing ceRNA network
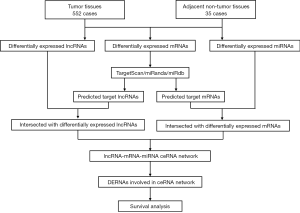
MiRcode (http://www.mircode.org/) was used to predict lncRNA-miRNA interactions (18). The mRNAs targeted by miRNAs were predicted cooperatively through TargetScan (http://www.TargetScan.org/mamm_31/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) and miRanda (http://www.Microrna.org/microrna/home.do) (19,20). To better understand how lncRNA mediates mRNA by combining with miRNA in UCEC, a network graph of ceRNA was constructed according to the above data and visualized utilizing Cytoscape v3.6.1. The flowchart of the ceRNA network construction is presented in Figure 1.
Survival analysis
To find the prognostic DERs signature, associating the clinical data of those patients with UCEC downloaded from TCGA, the survival curves of those samples with differentially expressed lncRNAs, miRNAs and mRNAs were drawn from using the “survival” package in R. Kaplan-Meier curve analysis was used to predict this univariate survival analysis. P values <0.05 was considered statistically significant.
Results
Identification of DERNAs
The Data Transfer Tool of the TCGA database was used to download RNAs expression profiles of UCEC patients and corresponding clinical information. We identified the significant DElncRNA, DEmRNAs and DEmiRNAs in UCEC samples compared with the normal samples using “edgeR” and limma in R language. As a result, a total of 1,102 lncRNAs, 2,612 mRNAs and 189 miRNAs were detected to be collectively dysregulated in UCEC. Among them, 314 and 788 lncRNAs were popularly upregulated and down-regulated, simultaneously, 1,641 and 971 mRNAs were collectively upregulated and down-regulated, severally. A total of 140 (74.07%) up-regulated and 49 (25.93%) down-regulated DEmiRNAs were identified (Figure 2). The heat map with complete link-age clustering of DElncRNAs, DEmRNAs and DEmiRNAs was built utilizing the “plots” package in R (data not shown).
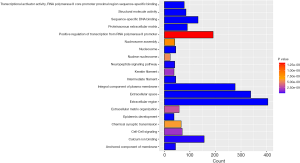
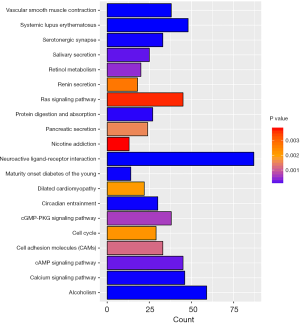
GO enrichment and KEGG pathway analyses
In order to further study of the functions of the differentially expressed genes (DEGs), we selected differentially expressed mRNAs for analysis. The 2,612 differentially expressed mRNAs were analyzed with DAVID bioinformatics resources. We select to show the top 20 GO biological processes and 20 KEGG pathways of the DEGs on account of the P-values (Figures 3,4). Among these pathways, the cAMP, GMP-PKG, Ras signaling pathways and CAMs were confirmed as UCEC-associated pathways.
ceRNA network in UCEC
For the sake of understand how lncRNA mediates mRNA through combining with miRNA in UCEC, we estimated the potential connections among the above corporately dysregulated genes according to the ceRNA hypothesis through computational analysis. We found that 27 key miRNAs were predicted to target 90 key lncRNAs based on miRcode (Table 1). The 27 key miRNAs were then used to predict key mRNAs using Targetscan, miRDB, and miRanda. Seventy-four mRNAs were found to interact with 20 miRNAs (Table 2). In the end, the ceRNA network that visualized based on the interactions among lncRNAs, miRNAs and mRNAs using Cytoscape v3.6.1 contained 90 lncRNAs, 27 miRNAs and 74 mRNAs (Figure 5).
Table 1
| lncRNA | miRNAs |
|---|---|
| AC002464.1 | hsa-mir-215, hsa-mir-383 |
| AC020907.1 | hsa-mir-195, hsa-mir-424, hsa-mir-216b |
| AC061975.6 | hsa-mir-106a |
| AC080037.1 | hsa-mir-429 |
| AC092422.1 | hsa-mir-195, hsa-mir-424, hsa-mir-383 |
| AC104472.1 | hsa-mir-143, hsa-mir-183, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-363, hsa-mir-425, hsa-mir-489 |
| AC107959.1 | hsa-mir-301b, hsa-mir-122, hsa-mir-383 |
| AC109462.1 | hsa-mir-301b, hsa-mir-143 |
| AC110491.1 | hsa-mir-140, hsa-mir-141, hsa-mir-200a, hsa-mir-143, hsa-mir-182, hsa-mir-215, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-216b, hsa-mir-508, hsa-mir-425, hsa-mir-489 |
| AC110619.1 | hsa-mir-140, hsa-mir-143, hsa-mir-184, hsa-mir-122 |
| AC112721.1 | hsa-mir-195, hsa-mir-424 |
| AC135178.1 | hsa-mir-106a, hsa-mir-122, hsa-mir-425, hsa-mir-489 |
| ADAMTS9-AS1 | hsa-mir-301b, hsa-mir-96, hsa-mir-145, hsa-mir-182 |
| ADAMTS9-AS2 | hsa-mir-301b, hsa-mir-106a, hsa-mir-96, hsa-mir-140, hsa-mir-141, hsa-mir-200a, hsa-mir-143, hsa-mir-145, hsa-mir-182, hsa-mir-183, hsa-mir-184, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-216b, hsa-mir-122, hsa-mir-363 |
| ADARB2-AS1 | hsa-mir-195, hsa-mir-424, hsa-mir-205, hsa-mir-216b |
| AGAP11 | hsa-mir-106a, hsa-mir-141, hsa-mir-200a, hsa-mir-143, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-182, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-216b |
| AL354707.1 | hsa-mir-216b |
| AL356133.2 | hsa-mir-106a, hsa-mir-383 |
| AL391152.1 | hsa-mir-143 |
| AL596188.1 | hsa-mir-205 |
| AL713998.1 | hsa-mir-141, hsa-mir-200a, hsa-mir-383 |
| ALDH1L1-AS2 | hsa-mir-301b, hsa-mir-106a, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-210, hsa-mir-508 |
| ARHGEF38-IT1 | hsa-mir-143, hsa-mir-425, hsa-mir-489 |
| ATXN8OS | hsa-mir-145, hsa-mir-183, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-210, hsa-mir-122 |
| BOK-AS1 | hsa-mir-184, hsa-mir-216b |
| BX255923.1 | hsa-mir-204, hsa-mir-211, hsa-mir-216b, hsa-mir-363 |
| C10orf91 | hsa-mir-106a, hsa-mir-140, hsa-mir-143, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-122 |
| C15orf54 | hsa-mir-301b, hsa-mir-106a, hsa-mir-143, hsa-mir-195, hsa-mir-424, hsa-mir-182, hsa-mir-429 |
| C2orf48 | hsa-mir-106a, hsa-mir-143, hsa-mir-195, hsa-mir-424, hsa-mir-183, hsa-mir-204, hsa-mir-211, hsa-mir-122 |
| C6orf99 | hsa-mir-140 |
| C8orf49 | hsa-mir-301b, hsa-mir-106a, hsa-mir-100, hsa-mir-143, hsa-mir-195, hsa-mir-424, hsa-mir-184, hsa-mir-429, hsa-mir-508, hsa-mir-122, hsa-mir-363 |
| DIO3OS | hsa-mir-106a, hsa-mir-143, hsa-mir-215, hsa-mir-204, hsa-mir-211, hsa-mir-508, hsa-mir-122, hsa-mir-383 |
| DLEU7-AS1 | hsa-mir-96, hsa-mir-195, hsa-mir-424, hsa-mir-182, hsa-mir-215 |
| DLX6-AS1 | hsa-mir-106a, hsa-mir-100, hsa-mir-141, hsa-mir-200a, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-215, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-216b, hsa-mir-122, hsa-mir-383 |
| DNM3OS | hsa-mir-301b, hsa-mir-145, hsa-mir-204, hsa-mir-211 |
| DSCR10 | hsa-mir-195, hsa-mir-424 |
| DSCR4 | hsa-mir-301b, hsa-mir-145, hsa-mir-216b, hsa-mir-508, hsa-mir-383, hsa-mir-425, hsa-mir-489 |
| DSCR4-IT1 | hsa-mir-204, hsa-mir-211, hsa-mir-508, hsa-mir-140, hsa-mir-143, hsa-mir-195, hsa-mir-424, hsa-mir-216b |
| DSCR8 | hsa-mir-205, hsa-mir-122 |
| ERVMER61-1 | hsa-mir-96, hsa-mir-182, hsa-mir-204, hsa-mir-211, hsa-mir-205 |
| FAM41C | hsa-mir-141, hsa-mir-200a, hsa-mir-145, hsa-mir-183, hsa-mir-205 |
| FRMD6-AS2 | hsa-mir-96, hsa-mir-143, hsa-mir-182, hsa-mir-184, hsa-mir-204, hsa-mir-211 |
| GLIS3-AS1 | hsa-mir-100, hsa-mir-140 |
| HAR1A | hsa-mir-216b, hsa-mir-122 |
| JAZF1-AS1 | hsa-mir-106a, hsa-mir-143, hsa-mir-205, hsa-mir-216b, hsa-mir-363 |
| KCNQ1DN | hsa-mir-145, hsa-mir-195, hsa-mir-424 |
| KIAA0087 | hsa-mir-96, hsa-mir-141, hsa-mir-200a, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-182, hsa-mir-183, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-383 |
| LINC00028 | hsa-mir-195, hsa-mir-424, hsa-mir-204, hsa-mir-211, hsa-mir-383 |
| LINC00051 | hsa-mir-140, hsa-mir-183, hsa-mir-205, hsa-mir-363 |
| LINC00052 | hsa-mir-145, hsa-mir-216b, hsa-mir-363, hsa-mir-425, hsa-mir-489 |
| LINC00113 | hsa-mir-145 |
| LINC00211 | hsa-mir-301b, hsa-mir-183, hsa-mir-184, hsa-mir-204, hsa-mir-211, hsa-mir-216b, hsa-mir-508, hsa-mir-383 |
| LINC00237 | hsa-mir-143, hsa-mir-205 |
| LINC00261 | hsa-mir-301b, hsa-mir-140, hsa-mir-143, hsa-mir-145, hsa-mir-182, hsa-mir-183, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-216b, hsa-mir-508 |
| LINC00314 | hsa-mir-106a, hsa-mir-143, hsa-mir-204, hsa-mir-211, hsa-mir-122 |
| LINC00336 | hsa-mir-96, hsa-mir-100, hsa-mir-143, hsa-mir-145, hsa-mir-216b |
| LINC00337 | hsa-mir-106a, hsa-mir-145, hsa-mir-182, hsa-mir-216b, hsa-mir-383, hsa-mir-143, hsa-mir-205 |
| LINC00355 | hsa-mir-140, hsa-mir-141, hsa-mir-200a, hsa-mir-195, hsa-mir-424, hsa-mir-122 |
| LINC00393 | hsa-mir-106a, hsa-mir-215 |
| LINC00443 | hsa-mir-301b, hsa-mir-141, hsa-mir-200a, hsa-mir-183, hsa-mir-204, hsa-mir-211, hsa-mir-363 |
| LINC00458 | hsa-mir-141, hsa-mir-200a, hsa-mir-143, hsa-mir-195, hsa-mir-424, hsa-mir-183, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-122, hsa-mir-383 |
| LINC00470 | hsa-mir-140, hsa-mir-141, hsa-mir-200a, hsa-mir-143, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-429, hsa-mir-216b, hsa-mir-508, hsa-mir-122, hsa-mir-383 |
| LINC00473 | hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-210 |
| LINC00488 | hsa-mir-96, hsa-mir-215, hsa-mir-205, hsa-mir-216b, hsa-mir-508, hsa-mir-122 |
| LINC00491 | hsa-mir-145, hsa-mir-184, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-216b, hsa-mir-122 |
| LINC00494 | hsa-mir-106a, hsa-mir-143, hsa-mir-145, hsa-mir-182, hsa-mir-210, hsa-mir-508, hsa-mir-383 |
| LINC00523 | hsa-mir-141, hsa-mir-200a, hsa-mir-215, hsa-mir-508, hsa-mir-122 |
| LSAMP-AS1 | hsa-mir-183 |
| MAGI2-AS3 | hsa-mir-106a, hsa-mir-141, hsa-mir-200a, hsa-mir-143, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-210, hsa-mir-216b, hsa-mir-508, hsa-mir-122, hsa-mir-363, hsa-mir-425, hsa-mir-489 |
| MEG3 | hsa-mir-301b, hsa-mir-106a, hsa-mir-96, hsa-mir-140, hsa-mir-141, hsa-mir-200a, hsa-mir-143, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-182, hsa-mir-184, hsa-mir-215, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-216b, hsa-mir-508, hsa-mir-122, hsa-mir-383 |
| MIR205HG | hsa-mir-301b, hsa-mir-143, hsa-mir-145, hsa-mir-183, hsa-mir-215, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-216b, hsa-mir-122, hsa-mir-383 |
| MIR210HG | hsa-mir-106a, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-182, hsa-mir-216b, hsa-mir-122 |
| MIR7-3HG | hsa-mir-145, hsa-mir-204, hsa-mir-211 |
| MYCNOS | hsa-mir-183, hsa-mir-205, hsa-mir-122, hsa-mir-363 |
| NAALADL2-AS2 | hsa-mir-183 |
| NAV2-AS2 | hsa-mir-96, hsa-mir-182, hsa-mir-204, hsa-mir-211, hsa-mir-122 |
| NKX2-1-AS1 | hsa-mir-301b, hsa-mir-96, hsa-mir-145, hsa-mir-182, hsa-mir-204, hsa-mir-211 |
| NOVA1-AS1 | hsa-mir-216b, hsa-mir-425, hsa-mir-489 |
| OXCT1-AS1 | hsa-mir-106a, hsa-mir-195, hsa-mir-424 |
| POU6F2-AS2 | hsa-mir-383 |
| PRICKLE2-AS1 | hsa-mir-106a, hsa-mir-141, hsa-mir-200a |
| PRICKLE2-AS2 | hsa-mir-145, hsa-mir-122 |
| RERG-IT1 | hsa-mir-182 |
| RMRP | hsa-mir-122 |
| RMST | hsa-mir-301b, hsa-mir-96, hsa-mir-145, hsa-mir-195, hsa-mir-424, hsa-mir-182, hsa-mir-429, hsa-mir-204, hsa-mir-211, hsa-mir-205, hsa-mir-508, hsa-mir-363, hsa-mir-425, hsa-mir-489 |
| SOX21-AS1 | hsa-mir-301b, hsa-mir-425, hsa-mir-489 |
| TBL1XR1-AS1 | hsa-mir-96, hsa-mir-182, hsa-mir-205 |
| TDRG1 | hsa-mir-215, hsa-mir-122 |
| TLR8-AS1 | hsa-mir-140, hsa-mir-182, hsa-mir-204, hsa-mir-211 |
| WARS2-IT1 | hsa-mir-140, hsa-mir-143, hsa-mir-195, hsa-mir-424, hsa-mir-216b |
miRNAs, microRNAs; UCEC, uterine corpus endometrial carcinoma; lncRNAs, long non-coding RNAs.
Table 2
| miRNA | mRNAs |
|---|---|
| hsa-mir-106a | FAM129A, KATNAL1, KIF23, RRM2, KPNA2, PLS1, ANKRD33B, CFL2HMGB3, E2F1, SALL3, TXNIP |
| hsa-mir-122 | AKT3, CLIC4 |
| hsa-mir-140 | PDGFRA |
| hsa-mir-141 | ZEB1, EPHA7, ZEB2 |
| hsa-mir-143 | IGFBP5, PAPPA |
| hsa-mir-145 | ABRACL, FLI1, DDC |
| hsa-mir-182 | ULBP2, TCEAL7, RECK, FOXF2, BDNF, NR3C1, MITF, CITED2, THBS1 |
| hsa-mir-183 | AKAP12, IDH2, NR3C1, ZEB1, CCNB1 |
| hsa-mir-195 | TGFBR3, WNT7A, AKT3, PSAT1, CEP55, RASSF2, RECK, KATNAL1, FGF2, ITPR1, E2F7, CBX2, RAB23, TPM2, KIF23, PRICKLE2, CLSPN, RUNX1T1, CCNE1, CDC25A |
| hsa-mir-200a | ZEB1, ZEB2, EPHA7 |
| hsa-mir-204 | ZCCHC24, ALPL, ITPR1, PRLR, CHRDL1 |
| hsa-mir-205 | LRRK2, ZEB1 |
| hsa-mir-210 | SERTM1 |
| hsa-mir-211 | POU3F2, ZCCHC24, PRLR, ITPR1, CHRDL1 |
| hsa-mir-216b | FZD5, MCM4 |
| hsa-mir-363 | ITPR1, S1PR1, TEF, SOX11, PAX9, SLC12A5 |
| hsa-mir-424 | E2F7, GNAL, RAB23, RASSF2, CLSPN, TGFBR3, CEP55, ITPR1, CBX2, CCNE1, RUNX1T1, TPM2, PSAT1, AKT3, RECK, KIF23, FGF2, CDC25A |
| hsa-mir-425 | MAP2K6 |
| hsa-mir-429 | ZFPM2, RASSF8, ZEB1, SHCBP1 |
| hsa-mir-96 | TRIB3, PROK2, ALK, ZEB1, JAZF1 |
miRNAs, microRNAs; UCEC, uterine corpus endometrial carcinoma; lncRNAs, long non-coding RNAs.
Survival analyses of DERs
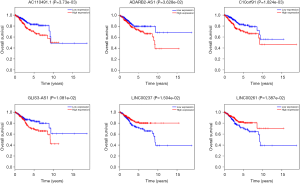
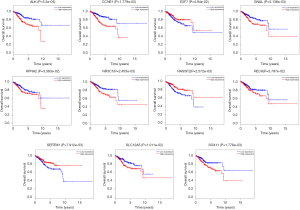
The overall survival for DERs in patients was researched to detect the relationship between differentially expressed RNAs (DERs) and the conditions of prognosis of patients with UCEC. Six of 90 DElncRNAs had a significant association with the overall survival of UCEC patients (P<0.05). As described in Figure 6, LINC00237 and LINC00261 had positive correlation with overall survival (P<0.05). AC11049.1, ADARB2-AS1, C10orf91 and GLIS3-AS1 were negatively correlated with overall survival (P<0.05). Three DEmiRNAs, has-mir-425, has-mir-211 and has-mir-301b were identified to be related to high levels of DEmiRNAs and with poor prognosis (Figure 7). Eleven DEmRNAs, including ALK, CCNE1, E2F7, GNAL, KPNA2, NR3C1, RASSF2, RECK, SERTM1, SLC12A5 and SOX11 were found to be associated with overall survival (Figure 8).

Discussion
In the last few years, cumulative evidence has indicated that lncRNAs play a vital part during tumor progression and may act as a molecular prognostic indicator and therapeutic target of tumors (21-23). lncRNAs function in many ways. Different studies have showed that the expression of aberrant lncRNAs presents a direction for diagnosis, prognosis, and therapy of different types of cancer (24).
The ceRNA hypothesis postulates that lncRNAs may compete with mRNAs for the binding sites of miRNAs and may further affect mRNA expression through MREs (5). The emergence of this hypothesis further complicates the relationship between miRNAs, mRNAs and lncRNAs and better explains the interaction among a variety of types of RNAs at the genetic level. Latest studies have confirmed that the differential expression of lncRNAs is compactly correlated with the pathogenesis and prognosis of tumors, and serve as a tumor-associated predictor (25,26). Zhang et al. demonstrated 7 lncRNAs had remarkable association with overall survival of rectal adenocarcinoma patients (6). Hu et al. accomplished lncRNA expression profiling in large colorectal cancer cohorts from Gene Expression Omnibus database and established a set included six lncRNAs that may be an effective means for evaluating clinical prognosis (27).
Although some researches have recognized several lncRNAs displaying upregulated and downregulated expression pattern in UCEC (28-32), these studies focused attention on recognizing differentially expressed lncRNAs. Of late, Zhou et al. (15) reported that 11 independent prognostic lncRNAs (RP11-1072A3.3.1, ACVR2B-AS1, RP4-781 K5.7.1, AC073046.25, AP001347.6, DOCK9-AS2, NRAV, GTF3C2-AS1, LINC01006, RP11-531A24.5 and AC004947.2) had a linear combination which was defined as an innovative lncRNA-focus expression signature to forecast survival of UCEC patients. To the best of our knowledge, no a whole genome-wide analysis of the lncRNA-miRNA-mRNA regulatory network of UCEC involved large number of samples has been reported. In this research, we confirmed UCEC-specific DElncRNAs, mRNAs and miRNAs based on the differential expression between tumor tissues and normal tissues. We successfully constructed lncRNA-miRNAs-mRNA ceRNA network and used it to disclose the interaction between specific lncRNAs and miRNAs and coding genes in UCEC. Compared with the routine samples, ninety DElncRNAs were recognized in UCEC samples. We evaluated the relationship between survival and all the DERs included in the ceRNA network. Finally, 6 lncRNAs, 3 miRNAs and 11 mRNAs were demonstrated to be remarkably associated with overall survival, which may provide meaningful guidance for the prospective research. The low expression of ADARB2-AS1 and high expression of LINC00261 in UCEC tissues may have the potential capacity to improve overall survival. It has been identified that ADARB2-AS1 has prognostic value for breast cancer (33). The decrease of lncRNA LINC00261 expression indicated a poor prognosis in gastric cancer and inhibited gastric cancer metastasis through impacting the transition of epithelial–mesenchymal (34).
We analyzed functions and pathways of UCEC-specific mRNAs using GO and KEGG. The GO enrichment analysis revealed that functions involved aspects of cellular functions and metabolism, etc. Several UCEC-associated pathways were confirmed by KEGG pathway analysis. The Ras signaling pathway is constantly varied in multiple different types of cancer, which is an early event driving tumorigenesis that has been observed and have also been connected with poor survival and treatment resistance (35). Saadatmand et al. (24) reported that CAMs play a significant part in the progression of metastasis. Previous studies have confirmed that wnt/β-catenin signaling can be inhibited by activating the cGMP/PKG pathway (36). These pathways and functions identified in our study may also be related to UCEC-specific lncRNAs due to altered interactions between lncRNAs and mRNAs in UCEC. We can further research the potential mechanism of the intersections among lncRNAs, mRNAs and miRNAs with the ceRNA network.
Several limitations should be taken into consideration in this research. In the present research, the ceRNA network was constructed by integrating bioinformatics analysis based on the hypothesis that lncRNAs can work as ceRNAs and regulate expression of the target genes through combining with miRNAs. One lncRNA could be in contact with a mass of miRNAs and concerned mRNAs indicated by the ceRNA network. Therefore, more research may concentrate on directing experiments to confirm several considerable interactions, specifically to validate the critical interactions to identify the effect of these lncRNAs as ceRNA in UCEC.
Conclusions
In conclusion, a lncRNA-associated ceRNA network was satisfactorily constructed, and it revealed the relationship among lncRNAs, miRNAs and mRNAs, and aided in elucidating the mechanisms underlying UCEC on the genetic level. In a great quantity of UCEC samples, six lncRNAs which were specific with cancer were recognized from generous candidates lncRNAs. Our study offers innovative DERs which can serve as candidates of prognosis biomarkers or potential therapeutic targets. Obviously, our results still necessitate to be further confirmed by prospective study in multicenter clinical study.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.44). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sponholtz TR, Palmer JR, Rosenberg L, et al. Reproductive factors and incidence of endometrial cancer in U.S. black women. Cancer Causes Control 2017;28:579-88. [Crossref] [PubMed]
- Cantrell LA, Backes F. Highlights from the Society of Gynecologic Oncology 2017 Annual Meeting on Women's Cancer. Gynecol Oncol 2017;145:483. [Crossref] [PubMed]
- Devis L, Moiola CP, Masia N, et al. Activated leukocyte cell adhesion molecule (ALCAM) is a marker of recurrence and promotes cell migration, invasion, and metastasis in early-stage endometrioid endometrial cancer. J Pathol 2017;241:475. [Crossref] [PubMed]
- Smogeli E, Davidson B, Cvancarova M, et al. L1CAM as a prognostic marker in stage I endometrial cancer: a validation study. BMC Cancer 2016;16:596. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Zhang Z, Wang S, Ji D, et al. Construction of a ceRNA network reveals potential lncRNA biomarkers in rectal adenocarcinoma. Oncol Rep 2018;39:2101-13. [PubMed]
- Xiong Y, Rong W, Peng L, et al. An integrated lncRNA, microRNA and mRNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 2017;8:85463-78. [Crossref] [PubMed]
- Li F, Huang C, Li Q, et al. Construction and Comprehensive Analysis for Dysregulated Long Non-Coding RNA (lncRNA)-Associated Competing Endogenous RNA (ceRNA) Network in Gastric Cancer. Med Sci Monit 2018;24:37-49. [Crossref] [PubMed]
- Zhao Y, Wang H, Wu C, et al. Construction and investigation of lncRNA-associated ceRNA regulatory network in papillary thyroid cancer. Oncol Rep 2018;39:1197-206. [PubMed]
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. [Crossref] [PubMed]
- Li S, Chen X, Liu X, et al. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol 2017;73:1-9. [Crossref] [PubMed]
- Hou P, Zhao Y, Li Z, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 2014;5:e1287. [Crossref] [PubMed]
- Liu X, Sun M, Nie F, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014;13:92. [Crossref] [PubMed]
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83. [Crossref] [PubMed]
- Zhou M, Zhang Z, Zhao H, et al. A novel lncRNA-focus expression signature for survival prediction in endometrial carcinoma. BMC Cancer 2018;18:39. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009;136:215. [Crossref] [PubMed]
- Nikolayeva O, Robinson MD. edgeR for Differential RNA-seq and ChIP-seq Analysis: An Application to Stem Cell Biology. Methods Mol Biol 2014;1150:45. [Crossref] [PubMed]
- Ashwini J, Marks DS, Erik L. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012;28:2062. [Crossref] [PubMed]
- RiffoCampos ÁL. Riquelme I, Brebimieville P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int J Mol Sci 2016;17:1987. [Crossref]
- Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res 2011;39:D163. [Crossref] [PubMed]
- Zhang L, Liang X, Li Y. Long non-coding RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and inactivating PI3K/AKT pathway. Oncol Rep 2017;38:2408. [Crossref] [PubMed]
- Yuan L, Huang S, Yan L, et al. Decreased expression of LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and EMT in colorectal cancer cells. Tumour Biol 2016;37:14205-15. [Crossref] [PubMed]
- Qiu JJ, Zhang XD, Tang XY, et al. ElncRNA1, a long non-coding RNA that is transcriptionally induced by oestrogen, promotes epithelial ovarian cancer cell proliferation. Int J Oncol 2017;51:507. [Crossref] [PubMed]
- Saadatmand S, de Kruijf EM, Sajet A, et al. Expression of cell adhesion molecules and prognosis in breast cancer. Br J Surg 2013;100:252-60. [Crossref] [PubMed]
- Augoff K, Mccue B, Plow EF, et al. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer 2012;11:5. [Crossref] [PubMed]
- Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014;5:2318-29. [PubMed]
- Hu Y, Chen HY, Yu CY, et al. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 2014;5:2230. [Crossref] [PubMed]
- Liu L, Chen X, Zhang Y, et al. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget 2017;8:31386-94. [PubMed]
- Yang X, Wang CC, Wyw L, et al. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett 2018;413:23. [Crossref] [PubMed]
- Maria AS, Marc D B, Hui L, et al. Long Non-Coding RNAs in Endometrial Carcinoma. Int J Mol Sci 2015;16:26463-72. [Crossref] [PubMed]
- Guo Q, Qian Z, Yan D, et al. LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by repressing Notch signaling. Biomed Pharmacother 2016;82:589-94. [Crossref] [PubMed]
- Guo C, Song WQ, Sun P, et al. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J Biomed Sci 2015;22:1-9. [Crossref] [PubMed]
- Yang F, Lyu S, Dong S, et al. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther 2016;9:761-72. [Crossref] [PubMed]
- Yu F, Wang YF, Su HF, et al. Decreased expression of the long noncoding RNA LINC00261 indicate poor prognosis in gastric cancer and suppress gastric cancer metastasis by affecting the epithelial-mesenchymal transition. J Hematol Oncol 2016;9:57. [Crossref] [PubMed]
- Caesar-Johnson SJ, Demchok JA, Felau I, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321. [Crossref] [PubMed]
- Li N, Xi Y, Tinsley HN, et al. Sulindac selectively inhibits colon tumor cell growth by activating the cGMP/PKG pathway to suppress Wnt/β-catenin signaling. Mol Cancer Ther 2013;12:1848-59. [Crossref] [PubMed]