
Serum amyloid A protein in cancer prognosis: a meta-analysis and systematic review
Introduction
Serum amyloid A protein (SAA), also known as serum amyloid protein A, is a 600-nucleotide long (1) and 122 amino acids protein and the encoding gene is located on chromosome 11p15.1 (2). In the human, SAA gene family contains the highly homologue SAA1, SAA2, SAA3 and SAA4 (3). SAA is an acute phase reactive protein as well as a high-density lipoprotein (HDL)-related protein (4,5). Meanwhile, SAA is expressed principally in hepar and extrahepatic cells or tissues such as epithelial cells, lymphocytes, and cancer cells (6) while occurring trauma, inflammation, infection, and oncogenesis (6,7).
SAA modulates cell adhesion, migration (8) in inflammation by inducing cytokines [IL-8 (9), G-CSF (10), etc.] expression and plays an important role in metabolism and transport of HDL and cholesterol (11). In addition, in the oncogenesis and tumor progression of various malignancies, chronic and lasting inflammation was considered as a crucial factor (12-15). But the function of SAA in oncogenesis still remains not clearly explained, and published studies suggest that SAA could become a biomarker to evaluate the growth of tumor and host response activity (16).
Following the explosive growth of knowledge of cancer biology, prognostic biomarker searching has been the most important fields of clinical oncology. A growing number of publications stated that chronic inflammation plays a prognostic role in many different cancers (17,18) and SAA status has been suggested as a prognostic biomarker for several tumors, such as renal cell carcinoma (RCC) (19), breast carcinoma (20), melanoma (21), lung cancer (22) and gastric carcinoma (23). The aim of this systematic review and meta-analysis was to clearly and comprehensively understand the prognostic value of SAA in different cancers with a standardized meta-analysis technique.
We present the following article in accordance with the PRISMA reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-3417).
Methods
Search strategy and study eligibility
Published studies were comprehensively searched on two independent databases PubMed and EBSCO till January 2020 using the MeSH words: “serum amyloid A protein” AND “Neoplasms”. SAA levels were determined by each author of the studies with immunity method value. Overall survival (OS), which is defined as the length of time that patients were still alive from initial cancer diagnosis, was the primary outcome. Prospective cohort studies and retrospective ones were both included. Articles were ruled out following the criteria: (I) non-abstract or non-English article; (II) case reports or review articles; (III) duplicate ones. The other respects of the studies like patient ethnicity, type and stage of tumor, length of follow up, and therapeutic schedule was not restricted (study eligibility criteria outlined in Table 1).
Table 1
| Study eligibility criteria | Detail |
|---|---|
| SAA definition | Serum amyloid a protein as defined by each study (no restriction regarding laboratory method, immunity method) |
| Outcome measure | Overall survival |
| Study design | Prospective or retrospective cohort studies |
| Patient age | Any |
| Anatomical site | Any |
| Study tissue | Any |
| Stage | Any |
| Therapy | Any |
| Length of follow-up | Any |
Publication identification and data extraction
To identify the eligibility of the studies, every publication was examined by two authors independently according to Meta-analysis Of Observational Studies in Epidemiology (MOOSE) guideline (24). To exclude the unrelated articles, the titles and abstracts of each study were read carefully with a unified standard. The remaining relevant studies were deeply examined to check their relevance. We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (25) to present the flow chart of study identification in Figure 1.
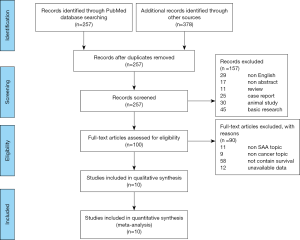
Two reviewers used the Newcastle-Ottawa Quality Assessment Scales to conduct the quality assessment independently for each study. And all differences between the two reviewers’ assessment were solved by consensus.
Statistical analysis
We used Review Manager 5 to conduct the statistical analysis. The patients were divided into two constitutive groups according to SAA levels (SAA-high, SAA-low) by each study. Heterogeneity of clinical factors and methodological sources between studies were taken into consideration to assess whether it is appropriate to complete such a statistical synthesis. We calculated the heterogeneity using Q test, and measured the discordance of pooled OR by I2 statistics (26). For this meta-analysis, we defined an I2 statistic of <50% as an indication of acceptable heterogeneity. Funnel plots were drew to evaluate the publication bias.
For the meta-analysis, OS was defined as the primary outcome. We used the weighted average of ORs to get a summary of the relationship between OS and SAA expression, which can partially correct the variance of OR in each publication. Mantel-Haenszel Fixed-effects (27) model was used to complete the synthesis when P<0.1, I2<50%, in the opposite, Random-effects (26) model used when P>0.1, I2>50%. In order to show the result of synthesis more clearly, forest plots presenting ORs of each study and pooled OR were completed. In the forest plots, the horizontal line segments indicated 95% confidence intervals (CIs), and the box in the middle of each line segment represented OR of each study. The vertical line was the reference line representing OR =1.0. At the bottom of the forest plots, the diamond represented the pooled OR, and the width represented the CIs. In order to evaluate the effect of cancer type of the result, subgroup analyses (RCC, lung cancer, digestive cancer, other or mixed) were conducted.
Results
Eligible studies
The 635 initially identified records were searched from two database (PubMed and EBSCO) with the MeSH words: “serum amyloid A protein” AND “Neoplasms” (flow chart of study identification summarized in Figure 1). Then, 424 records were excluded for the reason of duplication, non-English and non-abstract. While conducting the initial title/abstract screen, we excluded 111 publications just because they were reviewed, case report, animal study or basic research; 100 studies remained for full-text inspection after title/abstract screening. The remained 100 records were carefully assessed for the eligible studies (Figure 1). During this procedure, 90 full-text articles were excluded for unavailable data or unrelated to the main topic (SAA, cancer and survival) and ten publications (21,28-36), published from 1986 to 2020, were qualified for the meta-analysis for the success to extract the necessary data.
Study characteristics
The characteristics of these ten publications were presented in Table 2. The total included patient number of all the publications was 1496, ranging from 58 to 379 per study (median =150). Four studies assessed RCC (21,28,30,36), two evaluated lung cancer (29,31), and one each assessed melanoma (21), esophageal carcinoma (33), gastric cancer (34), and different cancer (35). Almost all studies included different disease stages except one study not reported such relevant information. However, one study (32) examined only stage IIb or IIIa cancers, one (31) examined only stage I and II cancers, and one (28) examined only stage IV cancers.
Table 2
| Study | Study characteristics | Patient population | SAA assessment | Outcome | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | Country (region) | Patient number | Type of tumor | Stage | Male gender, (N, %) | Median age (range) | Lab method | SAA classification | Definition of SAA (+) | SAA (+) prevalence, (N, %) | NOS score | OS/PFS | ||||
| Findeisen P, et al. (21) | 2009 | Germany | 379 | Melanoma | I-IV | 208, 54.9% | 54.7 (6.8–87.8) | Immunonephelometric assay | Low/high | SAA >10 mg/L | 203, 53.6% | 8 | OS | |||
| Vermaat JS, et al. (I) (28) | 2012 | The Netherlands | 114 | mRCC | IV | 76, 67% | 60 (49.9–70.1) | Conventional antibody-directed enumeration assays | Low/high | SAA >71 ng/mL | 52, 45.6% | 8 | PFS, OS | |||
| Vermaat JS, et al. (II) (28) | 2012 | The Netherlands | 151 | mRCC | IV | 107, 71% | 60.2 (50.1–70.3) | Conventional antibody-directed enumeration assays | Low/high | SAA >72 ng/mL | 82, 54.3% | PFS, OS | ||||
| Paret C, et al. (29) | 2010 | Germany | 87 | RCC | NR | NR | NR | Immunohistochemistry assay | Negative/positive | NR | 15, 17.2% | 7 | OS | |||
| Liu YS, et al. (30) | 2012 | China | 106 | Lung cancer | IIb, IIIa | NR | NR [41–73] | NR | Low/high | SAA >17.05 | 56, 52.8% | 7 | OS, RFS | |||
| Wang JY, et al. (33) | 2012 | China | 165 | Esophageal squamous cell carcinoma | I-IV | 129, 78.2% | 58.5 [40–70] | Poly-clonal antibody | Non-elevated/elevated | SAA >8.0 mg/L | 91, 55.2% | 7 | OS | |||
| Kwon HC, et al. (34) | 2012 | South Korea | 115 | Gastric cancer | I-IV | 68, 59.1% | NR | Latex agglutination turbidoimmunometric assays | Low/high | SAA >4.2 mg/mL | 57, 49.6% | 8 | DFS, OS | |||
| Biran H, et al. (35) | 1986 | Israel | 151 | Cancer | I-IV | 48, 31.8% | NR | Radioimmunoassay | Low/high | SAA >10 µg/mL | 39, 25.8% | 5 | OS | |||
| Kimura M, et al. (31) | 2001 | Japan | 72 | RCC | I-II | NR | 60.4 [34–81] | Latex agglutination turbidimetric immunoassay | Normal/elevated | SAA >8 mg/L | 38, 52.8% | 6 | OS | |||
| Ramankulov A, et al. (36) | 2008 | Germany | 98 | RCC | I-IV | 66, 67.3% | 60.0 [55–65] | ELISA | Low/high | SAA >27.7 mg/L | 30, 30.6% | 8 | OS | |||
| Wang YS, et al. (32) | 2013 | Taiwan | 58 | Lung cancer | I-IV | 42, 72.4% | 53.4% <70 years | ELISA | Low/high | SAA >670 µg/mL | 15, 25.9% | 8 | OS | |||
mRCC, metastatic renal cell carcinoma; RCC, renal cell carcinoma; SAA, serum amyloid A protein; PFS, progression-free survival; OS, overall survival.
Heterogeneity in SAA definition
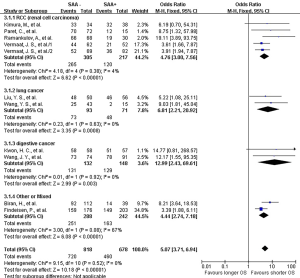
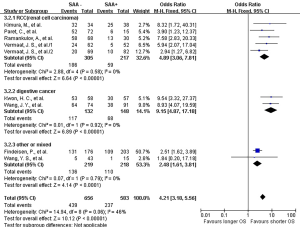
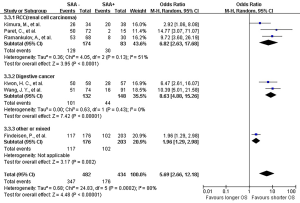
The lab methods extracted from each included studies were presented in Table 2. Different lab methods were used for evaluating the SAA status, including two studies (32,36) used ELISA, two (31,34) LATI (latex agglutination turbid metric immunoassay), two (21,29) immunohistochemistry assay, one (28) conventional antibody-directed enumeration assays, one (35) used radioimmunoassay, one (33) poly-clonal antibody and another one (30) was not reported. Different method and the staining score decided the different cutoff for elevated SAA status in each study. The total mean percentage of elevated SAA status patients was 45.3%. Melanoma and ESCC showed the highest percentage (far over 50%) of elevated SAA among the patients. The percentage of SAA overexpression in lung, gastric and renal cancer ranged 17.25% to 54.3%. Significant statistical heterogeneity was absent in 1-year survival (Q=9.15, I2=0%) (Figure 2), 3-year survival (Q=14.94, I2=46%) (Figure 3), but present in 5-year survival (Q=24.83, I2=80%) (Figure 4).
Meta-analysis
Total OS
The combined analysis for the included ten studies showed that SAA status had a strong correlation with a shorter OS (1-year survival OR for death =5.07, 95% CI, 3.71–6.94, P<0.00001) (Figure 2), while no study indicated that SAA was correlated with a longer OS. We can also draw a similar conclusion for the 3-year OS (OR for death =4.21, 95% CI, 3.18 to 5.56, P<0.00001) (Figure 3) and 5-year OS (OR for death =5.69, 95% CI, 2.66–12.18, P<0.00001) (Figure 4).
OS for different cancers
In renal carcinoma (21,28,30,36), a high SAA status showed an unfavorable OS (OR for death at 1 year =4.76, 95% CI, 3.00–7.56; OR for death at 3 years = 4.89, 95% CI, 3.06–7.81; OR for death at 5 years =6.82, 95% CI, 2.63–17.68) (Figures 2-4). The two studies of digestive carcinoma (33,34) indicated that elevated SAA status correlated with a worse OS than that in RCC (OR for death at 1 years =12.99, 95% CI, 2.43–69.61, P<0.0001; OR for death at 3 years =9.15, 95% CI, 4.87–17.18, P<0.00001; OR for death at 5 years =8.63, 95% CI, 4.88–15.26, P<0.00001) (Figures 2-4). However, there was no statistical difference between digestive and non-digestive cancers. High status of SAA reported in another two studies of lung cancer was also associated with an unfavorable 1-year OS (OR for death =6.81, 95% CI, 2.21–20.92) (Figure 2).
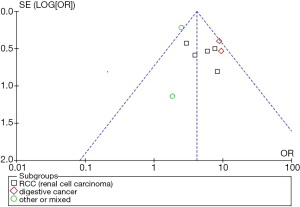
Publication bias
Publication bias analysis of the ten studies were presented as funnel plots (Figure 5) and the results indicate negligible publication bias
Discussion
A systematic review was carried out to evaluate the effect of SAA status on OS of patients with different kind of tumors. This meta-analysis showed a remarkable role of SAA in diverse cancer prognosis as well as in the subgroup of RCC, lung cancer and digestive carcinoma. No matter in early and locally advanced diseases (stages I–III) or in metastasis carcinoma (stage IV), our analysis showed an essential effect. Therefore, SAA could be included as one of the prognostic biomarker factors for cancers especially for RCC and digestive carcinoma. And should be selected with or for a further prospective study with our previous results CD 166 and CD 133 expressions.
Plenty of prognostic factors for different cancers existed in current references, such as breast cancer (37,38), bladder cancer (39,40), RCC (41), colorectal cancer (42), lung cancer and so on. The researchers have proved some clinical prognostic factors, mainly tumor staging and performance status (PS) (43). However, some clinical prognostic factors are not homogeneous, so that clinical oncologists cannot easily predict the survival for individual patients, such as tumor staging. Consequently, meta-analyses to evaluate the prognostic value of a new biological marker are urgent needed to solve the limitation statistical power of small size studies and to indicate more new prognostic factors, as our group published these years (44,45).
Heterogeneity is a significant problem that can influence the result of meta-analysis. Therefore, to avoid selection biases, we carried a methodological assessment as we implemented in prior studies (44,45). There was no obvious difference between the 10 eligible studies. However, it was hard to perform credible statistical comparisons with a limited study number of each subgroup.
Several limitations of our meta-analysis need to be taken into account while discussing our results. Primarily, potential confounding factors would not be corrected for the reason of the unavailable individual patient data such as age, gender or TNM. Such a literature-based analysis was unlikely to gain adequate information. Secondly, publication bias, selection bias, difference in lab method of detecting SAA and definition of SAA (+) between each study, and method of extrapolation of OR, were also factors that would directly influence our result.
In conclusion, our meta-analysis indicates that high SAA status is correlated with an unfavorable OS in different cancers, especially in RCC, and digestive cancer. Our present and prior results from meta-analyses serve to carry out a prospective multivariate trial aiming to observe which combination of classical and new prognostic biomarkers or which biomarkers will play a more important role in the prognosis of cancer patients.
Acknowledgments
Funding: This work was funded by
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-3417
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3417). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- De Buck M, Gouwy M, Wang JM, et al. Structure and Expression of Different Serum Amyloid A (SAA) Variants and their Concentration-Dependent Functions During Host Insults. Curr Med Chem 2016;23:1725-55. [Crossref] [PubMed]
- Taylor TD, Noguchi H, Totoki Y, et al. Human chromosome 11 DNA sequence and analysis including novel gene identification. Nature 2006;440:497-500. [Crossref] [PubMed]
- Sack GH Jr. Serum amyloid A - a review. Mol Med 2018;24:46. [Crossref] [PubMed]
- Sato M, Ohkawa R, Yoshimoto A, et al. Effects of serum amyloid A on the structure and antioxidant ability of high-density lipoprotein. Biosci Rep 2016;36:e00369. [Crossref] [PubMed]
- Coetzee GA, Strachan AF, van der Westhuyzen DR, et al. Serum amyloid A-containing human high density lipoprotein 3. Density, size, and apolipoprotein composition. J Biol Chem 1986;261:9644-51. [Crossref] [PubMed]
- Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol 2000;7:64-9. [Crossref] [PubMed]
- Rienhoff HY Jr, Huang JH, Li XX, et al. Molecular and cellular biology of serum amyloid A. Mol Biol Med 1990;7:287-98. [PubMed]
- Sozzani S, Del Prete A. Chemokines as relay signals in human dendritic cell migration: serum amyloid A kicks off chemotaxis. Eur J Immunol 2015;45:40-3. [Crossref] [PubMed]
- De Buck M, Berghmans N, Pörtner N, et al. Serum amyloid A1α induces paracrine IL-8/CXCL8 via TLR2 and directly synergizes with this chemokine via CXCR2 and formyl peptide receptor 2 to recruit neutrophils. J Leukoc Biol 2015;98:1049-60. [Crossref] [PubMed]
- Kim JC, Jung YS, Lee HY, et al. Serum amyloid A inhibits dendritic cell differentiation by suppressing GM-CSF receptor expression and signaling. Exp Mol Med 2017;49:e369. [Crossref] [PubMed]
- Shen C, Sun XG, Liu N, et al. Increased serum amyloid A and its association with autoantibodies, acute phase reactants and disease activity in patients with rheumatoid arthritis. Mol Med Rep 2015;11:1528-34. [Crossref] [PubMed]
- Zhong Z, Sanchez-Lopez E, Karin M. Autophagy, Inflammation, and Immunity: A Troika Governing Cancer and Its Treatment. Cell 2016;166:288-98. [Crossref] [PubMed]
- Schottenfeld D, Beebe-Dimmer J. Chronic inflammation: a common and important factor in the pathogenesis of neoplasia. CA Cancer J Clin 2006;56:69-83. [Crossref] [PubMed]
- Sfanos KS, Yegnasubramanian S, Nelson WG, et al. The inflammatory microenvironment and microbiome in prostate cancer development. Nat Rev Urol 2018;15:11-24. [Crossref] [PubMed]
- Nishizawa T, Suzuki H. Gastric Carcinogenesis and Underlying Molecular Mechanisms: Helicobacter pylori and Novel Targeted Therapy. Biomed Res Int 2015;2015:794378. [Crossref] [PubMed]
- Ren P, Sun D, Xin D, et al. Serum amyloid A promotes osteosarcoma invasion via upregulating αvβ3 integrin. Mol Med Rep 2014;10:3106-12. [Crossref] [PubMed]
- Kim WR, Han YD, Min BS. C-Reactive Protein Level Predicts Survival Outcomes in Rectal Cancer Patients Undergoing Total Mesorectal Excision After Preoperative Chemoradiation Therapy. Ann Surg Oncol 2018;25:3898-905. [Crossref] [PubMed]
- Kobayashi S, Matsumura Y, Karube Y, et al. Inflammation-Based Prognostic Score Predicts Postoperative Survival of Patients with Interstitial Pneumonia After Undergoing Lung Cancer Resection. World J Surg 2018;42:2143-52. [Crossref] [PubMed]
- Oz Atalay F, Aytac Vuruskan B, Vuruskan H. Significance of amyloid A immunoexpression in the prognosis of renal cell carcinoma. APMIS 2016;124:257-62. [Crossref] [PubMed]
- Pierce BL, Ballard-Barbash R, Bernstein L, et al. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J Clin Oncol 2009;27:3437-44. [Crossref] [PubMed]
- Findeisen P, Zapatka M, Peccerella T, et al. Serum amyloid A as a prognostic marker in melanoma identified by proteomic profiling. J Clin Oncol 2009;27:2199-208. [Crossref] [PubMed]
- Kanoh Y, Abe T, Masuda N, et al. Progression of non-small cell lung cancer: diagnostic and prognostic utility of matrix metalloproteinase-2, C-reactive protein and serum amyloid A. Oncol Rep 2013;29:469-73. [Crossref] [PubMed]
- Chan DC, Chen CJ, Chu HC, et al. Evaluation of serum amyloid A as a biomarker for gastric cancer. Ann Surg Oncol 2007;14:84-93. [Crossref] [PubMed]
- Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000;283:2008-12. [Crossref] [PubMed]
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 2010;8:336-41. [Crossref] [PubMed]
- Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [Crossref] [PubMed]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. [Crossref] [PubMed]
- Vermaat JS, Gerritse FL, van der Veldt AA, et al. Validation of serum amyloid alpha as an independent biomarker for progression-free and overall survival in metastatic renal cell cancer patients. Eur Urol 2012;62:685-95. [Crossref] [PubMed]
- Paret C, Schon Z, Szponar A, et al. Inflammatory protein serum amyloid A1 marks a subset of conventional renal cell carcinomas with fatal outcome. Eur Urol 2010;57:859-66. [Crossref] [PubMed]
- Liu YS, Luo XY, Li QR, et al. Shotgun and targeted proteomics reveal that pre-surgery serum levels of LRG1, SAA, and C4BP may refine prognosis of resected squamous cell lung cancer. J Mol Cell Biol 2012;4:344-7. [Crossref] [PubMed]
- Kimura M, Tomita Y, Imai T, et al. Significance of serum amyloid A on the prognosis in patients with renal cell carcinoma. Cancer 2001;92:2072-5. [Crossref] [PubMed]
- Wang YS, Chang HJ, Chang YC, et al. Serum amyloid a as a predictive marker for radiation pneumonitis in lung cancer patients. Int J Radiat Oncol Biol Phys 2013;85:791-7. [Crossref] [PubMed]
- Wang JY, Zheng YZ, Yang J, et al. Elevated levels of serum amyloid A indicate poor prognosis in patients with esophageal squamous cell carcinoma. BMC Cancer 2012;12:365. [Crossref] [PubMed]
- Kwon HC, Kim SH, Oh SY, et al. Clinicopathologic significance of expression of nuclear factor-kappaB RelA and its target gene products in gastric cancer patients. World J Gastroenterol 2012;18:4744-50. [Crossref] [PubMed]
- Biran H, Friedman N, Neumann L, et al. Serum amyloid A (SAA) variations in patients with cancer: correlation with disease activity, stage, primary site, and prognosis. J Clin Pathol 1986;39:794-7. [Crossref] [PubMed]
- Ramankulov A, Lein M, Johannsen M, et al. Serum amyloid A as indicator of distant metastases but not as early tumor marker in patients with renal cell carcinoma. Cancer Lett 2008;269:85-92. [Crossref] [PubMed]
- Guldberg TL, Christensen S, Zachariae R, et al. Prognostic factors in early breast cancer associated with body mass index, physical functioning, physical activity, and comorbidity: data from a nationwide Danish cohort. Breast Cancer Res Treat 2017;162:159-67. [Crossref] [PubMed]
- Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia 2001;6:375-92. [Crossref] [PubMed]
- Hernández-Fernández C, Herranz-Amo F, Moralejo-Gárate M, et al. Infiltrating bladder cancer: prognostic factors, follow-up and treatment of relapses. Actas Urol Esp 2017;41:352-8. [PubMed]
- Schalken JA. Molecular prognostic factors in bladder cancer. Eur Urol 1998;33:13-4. [Crossref] [PubMed]
- Motzer RJ, Bacik J, Mazumdar M. Prognostic factors for survival of patients with stage IV renal cell carcinoma: memorial sloan-kettering cancer center experience. Clin Cancer Res 2004;10:6302S-3S. [Crossref] [PubMed]
- Russo A, Bazan V, Agnese V, et al. Prognostic and clinicopathological role of high Ki-67 expression in patients with renal cell carcinoma: a systematic review and meta-analysis. Ann Oncol 2005;16:iv44-49. [Crossref] [PubMed]
- Paesmans M, Sculier JP, Libert P, et al. Prognostic factors for survival in advanced non-small-cell lung cancer: univariate and multivariate analyses including recursive partitioning and amalgamation algorithms in 1,052 patients. The European Lung Cancer Working Party. J Clin Oncol 1995;13:1221-30. [Crossref] [PubMed]
- Wang K, Xu J, Zhang J, et al. Prognostic role of CD133 expression in colorectal cancer: a meta-analysis. BMC Cancer 2012;12:573. [Crossref] [PubMed]
- Ni C, Zhang Z, Zhu X, et al. Prognostic value of CD166 expression in cancers of the digestive system: a systematic review and meta-analysis. PLoS One 2013;8:e70958. [Crossref] [PubMed]


