
Association between insulin-like growth factor 1 gene rs5742612 polymorphism and malignant tumor susceptibility: a meta-analysis
Introduction
Malignant tumors are currently one of the most serious diseases that endanger human health. There is an inseparable relationship between environment, gene, and malignant tumors (1). To some extent, a malignant tumor is a hereditary disease, but environmental factors and other non-hereditary factors also play a significant role in many stages of tumor genesis and development (2). It is now believed that genetic factors alone can only explain a small part of the pathogenesis of tumors, while the pathogenesis of most tumors is attributed to the synergistic effect between external environmental factors and the susceptibility to malignant tumors (3). It is worth noting that some of the susceptibility to malignant tumors comes from the diversity of the human genome, and related factors include pollution, smoking, drinking, overweight and infection (4).
In recent years, more and more attention has been paid to the role of insulin-like growth factor 1 (IGF1) in the genesis and development of malignant tumors (5). IGF1 is a growth factor involved in a large amount of significant biological and pathological processes (6). The important function of IGF1 is to promote cell proliferation and inhibit cell apoptosis, which is involved in the development of malignant tumors (7). Human IGF1 is just a little molecule of single-chain polypeptide composed of 70 amino acids (8), and its coding gene is located in L2Q22-24. It is an essential mitotic factor in vivo, which can promote cell proliferation and inhibit cell apoptosis. It often acts in autocrine and paracrine ways and controls the process of proliferation, differentiation, and apoptosis of various cells. In normal cells, IGF1 is not required, but in malignant cells, IGF1 has strong mitogenicity and anti-apoptotic activity (9). IGF1 can promote the differentiation and growth of tumor cells by secreting or paracrine tumor cells. Exogenous as a growth factor can also promote the growth of tumor cells. It makes up its receptor and activates phosphatidyl-nositol3-kinase (PI3K/Akt) signaling pathway and mitogen-activated protein kinase (MAPK) signaling pathway inhibiting tumor cell apoptosis and promote cell proliferation, respectively (10). Single nucleotide polymorphism (SNP) is an important part of individual genetic variation. IGF1 SNP associated with malignant tumor risk may serve as an important biomarker for the diagnosis of a malignant tumor (8-10). The study of twins shows that 40–60% of the individual variation of circulating IGF1 level depends on genetic factors, and the level of circulating IGF-1 is greatly influenced by SNP. Studies have shown that some IGF1 SNPs influence IGF1 levels in plasma, which affect the risk of the malignant tumor.
rs5742612 is located in the promoter region of the IGF1 gene, which may lead to selective splicing, which in turn leads to changes in protein function (11). It is reported that the substitution of G to C at the rs5742612 site leads to an increase in plasma IGF1 levels, which increases the risk of a malignant tumor (12).
Many epidemiological pieces of research have shown that SNPs of the IGF1 gene may be associated with malignant tumor risk (13). In the study of several common IGF1 gene polymorphisms, rs5742612 attracted much attention (12-14). However, studies on the relationship between rs5742612 and malignant tumor susceptibility are inconsistent. For example, Wong et al. (15) reported that rs5742612 was associated with the clinical prognosis of colorectal malignant tumor, but Mahmoudi et al. (16) showed that rs5742612 was not related to the susceptibility of the colorectal malignant tumor.
Given the differences between these studies, this study performed a meta-analysis of the association between rs5742612 and malignant tumor susceptibility. In a meta-analysis of 7 included articles, the results showed that the correlation between rs5742612 polymorphism of IGF1 gene and malignant tumor susceptibility was not significant. We present the following article in accordance with the PRISMA reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2005).
Methods
Search strategy
The study searched for related research in five databases: PubMed, Embase, Web of Science, CNKI, and Wanfang. Search conditions limit language to English and Chinese, publication data before January 25, 2020. Use the following keywords: “IGF1 or insulin-like growth factor 1 or IGF-1 or IGF or rs5742612”, “cancer or malignant tumor or tumor or carcinoma”, and “SNP or single nucleotide polymorphism or polymorphism or mutation”. The study also examined the references to the identified articles to make sure that we have access to all possibly relevant studies.
Inclusion and exclusion criteria
The inclusion criteria for this meta-analysis are as follows: (I) focus on the relationship between rs5742612 and malignant tumor susceptibility; (II) a case-control or cohort studies; (III) provide sufficient genotyping data to summarize the results (the genotype frequencies of TT, TC, and CC in case group and control group can be provided directly or calculated according to the data provided). The exclusion criteria are as follows: (I) exclude subjects with fewer subjects when subjects from both studies overlap; (II) exclude meta-analysis.
Data extraction
The two researchers extracted the following information from the included studies: first author name, year of publication, country, type of malignant tumor, ethnicity, genotyping method, control source, genotype distribution in case and control groups, and Hardy-Weinberg equilibrium.
Quality score
The quality of inclusion in RCT was evaluated from case source, control source, samples used to determine genotype, Hardy-Weinberg balance in the control group, and total sample size (Table 1). The full score is 15 points. The Newcastle Ottawa scale (NOS) was used to evaluate the methodological quality independently by two researchers. The differences are resolved through debate before an agreement is reached. Studies with high methodological quality should have a score of 5 or above.
Table 1
| Criterion | Score | |
|---|---|---|
| A | Case source | |
| Selected from the population or malignant tumor registry | 3 | |
| Selected from the hospital | 2 | |
| Selected from the pathology file but without description | 1 | |
| Without description | 0 | |
| B | Control source | |
| Based on population | 3 | |
| Blood donor or volunteer | 2 | |
| Hospital (no malignant tumor patients) | 1 | |
| Without description | 0 | |
| C | Sample for determining genotype | |
| White blood cell or normal tissue | 3 | |
| Tumor tissue or tissue exfoliation cells | 2 | |
| Without description | 0 | |
| D | Hardy-Weinberg equilibrium in the control group | |
| Balance | 3 | |
| Imbalance | 0 | |
| E | Total sample size | |
| ≥1,000 | 3 | |
| ≥500 and <1,000 | 2 | |
| ≥200 and <500 | 1 | |
| <200 | 0 |
Statistical analyses
RevMan software (version 5.3; Cochrane, London, UK) was used for statistical analysis. P value, odds ratio (OR), and 95% confidence interval (CI) were used to evaluate the correlation. For rs5742612, allele comparison (T vs. C) and codominant genotype comparison (TT vs. CC and TC vs. CC), dominant model (TT + TC vs. CC), and recessive model (TT vs. TC + CC) were examined. The heterogeneity was evaluated by inconsistent index I2 and chi-square test. The fixed-effects model can be employed to evaluate the set when the I2 value is less than 50%, which indicates that the heterogeneity is not statistically significant; otherwise, the random-effects model can be employed. The published bias was examined by Begg’s test.
Results
Search results and patient characteristics
After eliminating duplicates, 2,080 related articles were obtained by database retrieval. Then, by filtering the title and summary, we excluded 1,947 articles and left 133 articles. After reading the full text, we finally identified 7 articles that met the inclusion criteria., including 2,581 cases and 2,445 controls (Figure 1). The characteristics of the identified 7 articles are shown in Table 2. In the seven studies, six were performed in the Asian population and one in the European population. Of these, 1 involved colorectal cancer, 1 involved breast cancer, 2 involved gastric cancer, 2 involved prostate cancer, and 1 involved osteosarcoma. The quality scores of 6 studies were not less than 12 points. The genotypic distribution of the cases and the control group were shown in Table 3.
Table 2
| Study | Country | Tumor | Genotyping | Quality | |||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | Score | ||||
| Henningson 2011 (11) | Sweden | Breast cancer | TaqMan | 3 | 3 | 3 | 3 | 1 | 13 |
| Mahmoudi 2015 (16) | Iran | Colorectal cancer | TaqMan | 2 | 2 | 3 | 3 | 2 | 12 |
| Qian 2014 (17) | China | Prostatic cancer | TaqMan | 3 | 3 | 3 | 3 | 2 | 14 |
| Dong 2016 (18) | China | Prostatic cancer | TaqMan | 2 | 1 | 3 | 3 | 3 | 12 |
| Farahani 2015 (19) | Iran | Gastric cancer | TaqMan | 3 | 3 | 3 | 0 | 1 | 10 |
| Oh 2016 (20) | Korea | Gastric cancer | TaqMan | 3 | 3 | 3 | 3 | 3 | 15 |
| Mao 2017 (21) | China | Osteosarcoma | TaqMan | 2 | 3 | 3 | 3 | 1 | 12 |
A, case source; B, control source; C, Sample for determining genotype; D, Hardy-Weinberg equilibrium in the control group; E, total sample size.
Table 3
| Study | Genotypic (n) | Hardy-Weinberg equilibrium | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case group | Control group | |||||||||
| Total | TT | TC | CC | Total | TT | TC | CC | |||
| Henningson 2011 (11) | 100 | 58 | 38 | 4 | 110 | 69 | 36 | 5 | 0.052 | |
| Mahmoudi 2015 (16) | 339 | 322 | 16 | 1 | 261 | 249 | 11 | 1 | 0.877 | |
| Qian 2014 (17) | 503 | 245 | 209 | 49 | 227 | 107 | 94 | 26 | 0.301 | |
| Dong 2016 (18) | 664 | 293 | 292 | 79 | 702 | 361 | 276 | 65 | 0.620 | |
| Farahani 2015 (19) | 234 | 134 | 97 | 3 | 272 | 131 | 138 | 3 | <0.01 | |
| Oh 2016 (20) | 568 | 378 | 169 | 21 | 698 | 452 | 209 | 37 | 0.673 | |
| Mao 2017 (21) | 173 | 76 | 79 | 18 | 175 | 83 | 76 | 16 | 0.113 | |
T, thymidine; C, cytimidine.
Association between rs5742612 and malignant tumor susceptibility
Table 4 shows the general results of the Meta-analysis. No statistically significant evidence showed that IGF1 (rs5742612) SNP was associated with malignant tumor susceptibility (T vs. C, OR =0.99, 95% CI: 0.85–1.15, P=0.91, Figure 2; TC vs.CC: OR =1.03, 95% CI: 0.81–1.32, P=0.79, Figure 3; TT vs. CC: OR =0.92, 95% CI: 0.73–1.17, P=0.52, Figure 4; TT + TC vs. CC: OR =0.97, 95% CI: 0.77–1.22, P=0.80, Figure 5; TT vs. TC + CC: OR =0.98, 95% CI: 0.81–1.18, P=0.83, Figure 6).
Table 4
| Study | OR (95% CI) | P | I2 (%) |
|---|---|---|---|
| Allelic model: T vs. C | 0.99 (0.85–1.15) | 0.91 | 51 |
| Heterozygote model: TC vs. CC | 1.03 (0.81–1.32) | 0.79 | 0 |
| Homozygous model: TT vs. CC | 0.92 (0.73–1.17) | 0.52 | 15 |
| Dominant model: TT + TC vs. CC | 0.97 (0.77–1.22) | 0.80 | 0 |
| Recessive model: TT vs. TC + CC | 0.98 (0.81–1.18) | 0.83 | 52 |
T, thymidine; C, cytimidine.
Sensitivity analysis
A leave-one analysis is employed to assess the sensitivity of the identified 7 articles in the meta-analysis. When any of the studies were deleted, the overall statistical significance was not reversed, which indicates that the results remained stable despite the inclusion or exclusion of Hardy-Weinberg imbalance control groups. Therefore, the analysis results are relatively stable and credible in the meta-analysis.
Publishing bias
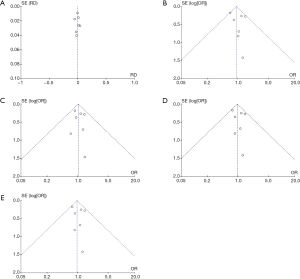
The publication deviation in the included study was evaluated by Begg’s test. No publication deviation was found in the current meta-analysis (Figure 7).
Discussion
The susceptibility to malignant tumors is influenced by many factors, such as genetic factors, environmental factors, hormone disorders, and infection (16). Although the exact mechanism of a malignant tumor is still unclear, the genetic factors such as IGF1 gene, estrogen receptor 1 gene and ladybug homologous box 1 gene are closely related to the occurrence and development of malignant tumor (17). Among them, IGF1, as a growth factor, has a wide range of physiological functions, which can control the proliferation, differentiation and apoptosis of various cells. The differential expression of the IGF1 gene may be involved in the development of malignant tumors (15-17).
Moreover, some researches have shown that the T/C polymorphism in the promoter region of the IGF1 gene may affect the level and functional activity of IGF1 protein (18). It is reported that many SNP is associated with the susceptibility to malignant tumors, so they may be an important biomarker for the diagnosis of malignant tumors potentially (19).
Many studies have confirmed that several IGF1 SNPs are associated with susceptibility to malignant tumors. This SNP includes rs6214, rs6220, rs35767, and rs5742612. It is reported that the substitution of G to C at the rs5742612 site leads to an increase in plasma IGF1 levels, which increases the risk of the malignant tumor (20). Since rs5742612 has a relatively small frequency of about 10–40% in the population included in the third phase of the Human 1000 Genome Project, so this study pays special attention to rs5742612 (21). Recently, related studies have explored the relationship between IGF1 rs5742612 SNP and the genesis and development of malignant tumors, but the results are not consistent. Leroy et al. (22) studied the genotyping of IGF1 gene rs5742612 SNP, which includes 87 malignant tumor cases and 87 nationalities and gender-matched control, healthy people. The results showed that there was a significant association between IGF1 gene rs5742612 SNP and malignant tumor susceptibility in the Chinese population. However, Mahmoudi et al. (16) showed that rs5742612 SNP was not associated with colorectal cancer susceptibility.
In this meta-analysis, we conducted a comprehensive and systematic search of the literature on IGF1 gene rs5742612 SNP and malignant tumors in English databases (Embase, PubMed, and Web of Science) and Chinese databases. After deleting the duplicate files, we initially obtained 2,080 related articles and ensured the maximum possible recall rate. Through a meta-analysis of the 7 identified articles, we found that the correlation between allele and genotype frequency of IGF1 gene rs5742612 polymorphism between malignant tumor cases and control healthy people was not statistically significant, which was consistent with the results of individual studies (19). However, these findings are inconsistent with the results reported by Qian et al. (17) and Dong et al. (18). Therefore, further research is needed on the role of rs5742612 in malignant tumor susceptibility risk.
Hardy-Weinberg’s principle is to detect whether the genotype and allele frequency observed in parent-child populations are in balance (23). Group stratification leads to hardy Weinberg bias, which may be a mixed association. Among the seven studies, only Farahani et al.’s study (19) is a Hardy-Weinberg imbalance, which may affect the overall results. However, after the exclusion of the study, there was no reversal in overall statistical significance. Therefore, this study is relatively stable, effective, and reliable. To ensure sufficient statistical ability, there must be a sufficient sample size. However, the relatively small sample size of the three studies in this meta-analysis may reduce the statistical strength of the test correlation, leading to debates on the results and impact on the conclusions (11,19,21). Compared with different races and populations, our meta-analysis showed that there was no significant correlation between IGF1rs5742612 polymorphism and malignant tumor susceptibility in Iran (16,19) or South Korea (20) population, which is inconsistent with the Chinese population (17,18,21). Due to the diversity and potential differences in malignant tumor susceptibility in the population, the results of the current meta-analysis may not be generalized, so further research is needed on other races and populations to provide additional evidence. Besides, the meta-analysis of this study has several limitations. First, the study did not consider potential external factors such as gender, age, diet, smoking and drinking habits, or genetic interactions. Second, the study only includes literature written in English and Chinese, and some important related research in other languages may be overlooked.
In summary, all existing studies that reported the association between IGF1 gene rs5742612 polymorphism and malignant tumor susceptibility were used in this meta-analysis. Ultimately this meta-analysis identified seven articles that meet the inclusion criteria, which includes 2,581 cases and 2,445 controls. P value, OR, and 95% CI were used to evaluate the correlation. For rs5742612, allele comparison (T vs. C) and codominant genotype comparison (TT vs. CC and TC vs. CC), dominant model (TT + TC vs. CC) and recessive model (TT vs. TC + CC) were examined. The heterogeneity was evaluated by inconsistent index I2 and chi-square test. The published bias was examined by Begg’s test. Table 4 shows the general results of the Meta-analysis. No statistically significant evidence showed that IGF1 (rs5742612) SNP was associated with malignant tumor susceptibility in the meta-analysis. However, due to the limited sample size included in the study, the only European study included 230 Swedish women, which may not represent the vast majority of “non-Asian” populations, further research should include different races, countries and larger sample size to ensure the accuracy and reliability of meta-analysis results and identify potential associations.
Acknowledgments
Funding: This article was funded by
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2005
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2005). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shen J, Liao Y, Hopper JL, et al. Dependence of cancer risk from environmental exposures on underlying genetic susceptibility: an illustration with polycyclic aromatic hydrocarbons and breast cancer. Br J Cancer 2017;116:1229-33. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A, et al. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Barrdahl M, Rudolph A, Hopper JL, et al. Gene-environment interactions involving functional variants: Results from the Breast Cancer Association Consortium. Int J Cancer 2017;141:1830-40. [Crossref] [PubMed]
- Nanda SS, Gandhi AK, Rastogi M, et al. Evaluation of XRCC1 Gene Polymorphism as a Biomarker in Head and Neck Cancer Patients Undergoing Chemoradiation Therapy. Int J Radiat Oncol Biol Phys 2018;101:593. [Crossref] [PubMed]
- Matsuoka H, Makimura C, Koyama A, et al. Prospective replication study implicates the catechol-O-methyltransferase Val158Met polymorphism as a biomarker for the response to morphine in patients with cancer. Biomed Rep 2017;7:380-4. [Crossref] [PubMed]
- Xu HY, Zhu DW, Zhong LP, et al. Effects of insulin-like growth factor binding protein 3 on cell growth and tumorigenesis in oral squamous cell carcinoma. Transl Cancer Res 2019;8:1709-17. [Crossref]
- Yao Y, Mao W, Dong M, et al. Serum Insulin-Like Growth Factor-1 (IGF-1): a Novel Prognostic Factor for Early Recurrence of Hepatocellular Carcinoma (HCC). Clin Lab 2017;63:261-70. [Crossref] [PubMed]
- Hendriks SH, Schrijnders D, Hateren KJV, et al. Association between body mass index and obesity-related cancer risk in men and women with type 2 diabetes in primary care in the Netherlands: a cohort study (ZODIAC-56). BMJ Open 2018;8:e018859. [Crossref] [PubMed]
- Kasprzak A, Kwasniewski W, Adamek A, et al. Insulin-like growth factor (IGF) axis in cancerogenesis. Mutat Res Rev Mutat Res 2017;772:78-104. [Crossref] [PubMed]
- Niu H, Shin H, Feng G, et al. Functional Single Nucleotide Polymorphism (SNP) Correlates With Clinical Outcome in Patients With Advanced Solid Tumors Treated With Alisertib, an Investigational Aurora A Kinase Inhibitor. Ebiomedicine 2017;25:50-7. [Crossref] [PubMed]
- Henningson M, Hietala M, Törngren T, et al. IGF1 htSNPs in relation to IGF-1 levels in young women from high-risk breast cancer families: implications for early-onset breast cancer. Fam Cancer 2011;10:173-85. [Crossref] [PubMed]
- Wang R, Xu D, Liu R, et al. Microsatellite and Single Nucleotide Polymorphisms in the Insulin-Like Growth Factor 1 Promoter with Insulin Sensitivity and Insulin Secretion. Med Sci Monit 2017;23:3722-36. [Crossref] [PubMed]
- Cao Y, Lindström S, Schumacher F, et al. Insulin-like Growth Factor Pathway Genetic Polymorphisms, Circulating IGF1 and IGFBP3, and Prostate Cancer Survival. J Natl Cancer Inst 2014;106:dju085. [Crossref] [PubMed]
- Milne E, Greenop KR, Metayer C, et al. Fetal growth and childhood acute lymphoblastic leukemia: Findings from the childhood leukemia international consortium. Int J Cancer 2013;133:2968-79. [Crossref] [PubMed]
- Wong HL, Koh WP, Probst-Hensch NM, et al. Insulin-like growth factor-1 promoter polymorphisms and colorectal cancer: a functional genomics approach. Gut 2008;57:1090-6. [Crossref] [PubMed]
- Mahmoudi T, Majidzadeh-A K, Karimi K, et al. An exon variant in insulin receptor gene is associated with susceptibility to colorectal cancer in women. Tumour Biol 2015;36:3709-15. [Crossref] [PubMed]
- Qian J, Zhou H, Chen J, et al. Genetic Polymorphisms in IGF-I and IGFBP-3 Are Associated with Prostate Cancer in the Chinese Population. PLoS One 2014;9:e85609. [Crossref] [PubMed]
- Dong YL, Zhang L, Zhang QJ, et al. Correlation between IGF-1 and IGFBp-3 gene polymorphism and prostate cancer susceptibility. Chinese Journal Of Surgical Oncology 2016;8:84-90.
- Farahani RK, Azimzadeh P, Rostami E, et al. Evaluation of insulin like growth facror-1 genetic polymorphism with gastric cancer susceptibility and clinicopathological features. Asian Pac J Cancer Prev 2015;16:4215-8. [Crossref] [PubMed]
- Oh SY, Shin A, Kim SG, et al. Relationship between insulin-like growth factor axis gene polymorphisms and clinical outcome in advanced gastric cancer patients treated with FOLFOX. Oncotarget 2016;7:31204-14. [Crossref] [PubMed]
- Mao J, Zhuang G, Chen Z. Genetic Polymorphisms of Insulin-Like Growth Factor 1 Are Associated with Osteosarcoma Risk and Prognosis. Med Sci Monit 2017;23:5892-8. [Crossref] [PubMed]
- LeRoy EC, Moore JH, Hu C, et al. Genes in the insulin and insulin-like growth factor pathway and odds of metachronous colorectal neoplasia. Hum Genet 2011;129:503-12. [Crossref] [PubMed]
- Ong J, Salomon J, te Morsche RH, et al. Polymorphisms in the Insulin-Like Growth Factor Axis Are Associated with Gastrointestinal Cancer. PLoS One 2014;9:e90916. [Crossref] [PubMed]








