
The E3 protein ubiquitin ligase Itch is a potential target in myeloid malignancies with marrow fibrosis
Introduction
BM fibrosis is characterized by the increased deposition of reticulin fibers or collagen. Myeloid malignancies like myelodysplastic syndrome (MDS) and acute myelocytic leukemia (AML) with BM fibrosis are associated with adverse prognosis (1-3). But the underlying mechanism of these diseases has not been fully understood yet. The gene expression profile of these kinds of diseases was not much reported either. But the molecular mechanism of MDS-F and AML-F may be similar to the primary myelofibrosis (PMF). The pathogenesis of PMF gets involved in many factors and transforming growth factor beta (TGF-β) is a classical signaling pathway in the pathogenesis of PMF (4). In this study we aimed to investigate the pathogenic mechanism of MDS-F and AML-F in order to get more knowledge of the diseases.
Itch is a monomeric protein, which belongs to the homologous to E6-AP carboxy terminus (HECT)-type family of E3 protein ubiquitin ligases. It plays key roles in different cellular contexts including DNA damage response, T-cell differentiation, the immune response, and cell death due to its functionally distinct substrates. Its deficiency in mice had been shown to cause severe autoimmune disease (5-11). Itch could also get involved in the TGF-β signaling (12) and may play a role in the generation of marrow fibrosis. The protein p73 participates in the apoptotic response to DNA damage and it is also one of the substrates of Itch (13). Deepa et al. found that Itch could downregulate p73 in the K562 cell line and the down-regulation of Itch might finally induce the apoptosis of the cells (14). So Itch may be a crucial protein in the pathogenesis of MDS-F and AML-F.
Elevated phosphatidylinositol 3-kinase (PI3K)/Akt oncogenic signaling is considered a hallmark of carcinomas as well as the myeloid malignancies (15,16). It was reported that the pathway could get involved in the regulation of Itch (17,18) but whether the PI3K/Akt pathway has a relationship with Itch in the myeloid malignancies hasn’t been much reported.
We first retrieved a dataset from the Gene Expression Omnibus (GEO) database about PMF which was then analyzed by bioinformatic methods to get some hints. We found that the E3 protein ubiquitin ligase Itch and the PI3K/Akt pathway proteins had differential expression in the PMF patients when compared with healthy controls. Then we used the K562 cell line which could express Itch protein and had activated PI3K/Akt pathway as a model to figure the relation between the pathway and Itch out. Finally, we assayed the Itch mRNA level in the clinical samples to have a knowledge of its role in the myeloid diseases preliminarily.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-3115).
Methods
Affymetrix microarray data
The microarray data of GSE53482 was retrieved and downloaded from the GEO (Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/) database based on the platform of GPL13667 Affymetrix Human Genome U219 Array. The total RNA of CD34+ cells from 42 PMF patients and 31 healthy donors were used in the study to develop the microarray data (Affymetrix, Inc., Santa Clara, CA, USA).
Differential expression analysis
We used the GEO2R analytical tool of the GEO database to perform the differential expression analysis of mRNA from the samples above. |Log2FC|>1 and adj. P value <0.01 were considered as cutoff values for the differentially expressed genes.
Protein-Protein interaction (PPI) network construction and sub-network identification
The Search Tool for the Retrieval of Interacting Genes (STRING) database is often used to perform the PPI network construction. In this study, we used this database to construct the PPI network of the differentially expressed genes. In the PPI network, each node stands for a gene and the edges stand for the interaction between the nodes. Degree is a parameter which can indicate the number of edges linked to a given gene node. We defined the hub gene of the network by “degree ≥15”. Only experiments and database were used as active interaction sources. Combined score >0.9 were thought as significant.
Gene ontology (GO) and pathways enrichment analyses
The Database for Annotation, Visualization and Integrated Discovery (DAVID) is often used to perform the functional annotation like GO and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database pathways enrichment analyses for the differentially expressed genes. In this study, we used this database to analyze the focused differentially expressed genes at the functional level.
Reagents and antibodies
Fetal bovine serum was purchased from NQBB International Biological Corporation (NQBB, Australia), Roswell Park Memorial Institute (RPMI)1640, penicillin and streptomycin were obtained from GENOM Inc. (Hangzhou, China). PI3K inhibitor LY294002 was purchased from Selleck Chemicals (Selleck, USA). Cell Counting Kit-8 (CCK-8) was product of DOJINDO LABORATORIES (DOJINDO, JAPAN), cell cycle and apoptosis testing kit was obtained from Invitrogen Inc. (Invitrogen, USA). Primers of mRNA ITCH (human, No. DHS905450), ACTB (human, No. DHS579991) were from Xing yuan biotechnology Co., Ltd (Shanghai, China). Antibodies for western blot were products of CST & Abcam. (β-Actin CST#4970, Akt CST#9272S, pAkt1/2/3 Abcam ab192623, P27 Abcam ab32034, Bcl-2 Abcam ab182858, Bax Abcam ab32503, Caspase-3 and cleaved Caspase-3 Abcam ab32351, Itch Abcam ab108515, P73 Abcam ab189896).
Cells and culture
Myelogenous leukemia cell line K562 (RRID:CVCL_0004) was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were placed at 37 °C in a humidified atmosphere with 5% CO2 and cultured with RPMI1640 media supplemented with 10% fetal bovine serum and with penicillin and streptomycin.
Cell proliferation assay with CCK-8
One hundred microlitre K562 cell suspension of logarithmic phase were plated per well in 96-well plates cultured with LY294002 20 µmol/L, 40 µmol/L or with no treatment for 48 h. The media without cells but added LY294002 with corresponding concentration was used as blank. The wells with cells without treated were control groups. Groups with LY294002 20 and 40 µmol/L were referred to as group C+20 and C+40 respectively. Then all the wells were added with 10 µL CCK-8 solution and incubated in the incubator for 4h. Finally, we used a microplate reader to measure the absorbance at 450 nm. ELX-808 Microplate spectrophotometer was product of VEDENG Technology Ltd. (Nanjing, China). The results of the groups were then statistically analyzed.
Apoptosis assay by Flow Cytometry
A total of 1×106 cells of each group mentioned above were harvested after the incubation period. Cells were washed with cold Phosphate Buffer Saline (PBS) twice after centrifugation with 1,200 rpm × 5 min, then the cells were re-centrifuged and discarded the supernatant. The cells were resuspended with 100 µL binding buffer in each tube. Adding 5 µL Annexin-V and 1 µL PI working buffer into each tube. After being gently vibrated, the cells were incubated in dark place at room temperature for 15 minutes. Adding 400 µL binding buffer into each tube, mixed them gently and kept the samples on ice. Finally, analyzing the stained cells by flow cytometry, measuring the apoptosis proportion of the cells in each tube. Flow cytometry was performed on a FACS Calibur (BD, USA).
Cell cycle assay by Flow Cytometry
Cells were harvested and washed as mentioned previously. Adding 4 °C pre-cooling 70% ethyl alcohol into each tube and then put them at 4 °C for 24 h. Discarded the supernatant after centrifugation with 1,200 rpm × 5 min and washed the cells with PBS twice. Added 500 µL PI/RNAse into each tube, mixed and incubated them at room temperature for 30 minutes. Flow Cytometry detected the cell cycle proportion of each group. Modfit 3.1 software analyzed the data and then the results were statistically analyzed.
RNA extraction and Real-time PCR
Extraction of total RNA from the cell line and clinical samples and the performance of Real-time PCR were all followed the direction of the TransZol Up Plus RNA Kit &Trans Script Green One-Step qRT-PCR Super Mix Kit (purchased from Trans Gen Biotech Inc., Beijing, China). Real-time PCR reaction system consisted of 5 µL RNA Template (1 pg–1 µg), 10 µL 2×TransScript® Tip Green qPCR Super Mix, 2 µL Forward/Reverse GSP (10 µM), 0.4 µL TransScript® One-Step RT/PI Enzyme Mix, RNase-free Water 2.6 µL. Reaction conditions: Reverse Transcription, 45 °C 5 min, then pre-incubation, 94 °C 30 s. 40 cycles amplification, 95 °C 10 s, 60 °C 30 s. ACTB as reference. Real-time PCR was performed on a Light Cycler® 480 (Roche Diagnostics Ltd). 2-ΔΔCt value of each cell line group mentioned above and 2-ΔCt value of the clinical samples were statistically analyzed.
Western blotting (WB)
Cells were collected from each group and eliminated the supernatant after centrifugation (1,200 g × 5 min), then the cells were lysed by the whole cell lysis buffer. The protein concentration of cell lysates was tested by Bicinchoninic Acid (BCA) assay. Then the lysates were heated in boiled water with loading buffer for 10–15 min. A total of 40 µg protein of each test was used for SDS-PAGE electrophoresis. Then proteins were transferred to nitrocellulose filter membrane (NC) membranes which were then saturated by Tris Buffer Saline (TBS) buffer containing 5% non-fat milk at room temperature for 1 h. Primary antibodies in TBS-Tween buffer with milk incubated with the membranes at 4 °C overnight. After three washes in TBS-Tween buffer, secondary antibodies incubated with the membranes at room temperature for 1h. Antibody binding was visualized using a chemiluminescent substrate. WB antibody binding was tested by Amersham Imager 1600. Quantification was done using Image J software. β-actin as reference, Experiments were repeated at least three timed and then statistically analyzed.
Clinical samples
Thirty-nine peripheral blood samples from 28 patients with MDS or AML (almost all converted from MDS) (Table 1) and 11 healthy subjects were collected and processed for RNA extraction. All the patients are de novo or didn’t get remission after treatment (the MDS patients were diagnosed with the WHO standard 2008 and IPSS was used as prognostic scoring system). This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of our hospital (approval number: 2020-089). Signed informed consents were obtained from all donators.
Table 1
| Variables | Range/N | Mean ± standard |
|---|---|---|
| Age (years old) | 25–82 | 61.46±13.68 |
| Gender | ||
| Male | 17 | |
| Female | 11 | |
| Diseases | ||
| MDS | 21 | |
| AML | 7 | |
| Blood routine examination | ||
| WBC (×109/L) | 0.3–49.8 | 10.57±14.01 |
| Hb (g/L) | 26–106 | 62.43±16.32 |
| PLT (×1012/L) | 1–384 | 69.89±83.97 |
| MDS IPSS classification | ||
| Low risk | 11 | |
| High risk | 8 | |
| WBC classification, ×109/L | ||
| ≤10 | 19 | |
| >10 | 9 | |
| Classification by BM fibrosis | ||
| With no BM fibrosis | 9 | |
| With BM fibrosis | 19 | |
| Classification by karyotype | ||
| Normal karyotype | 12 | |
| Abnormal karyotype | 13 |
MDS, myelodysplastic syndrome; AML, acute myelocytic leukemia; Hb, hemoglobin value; PLT, platelet count; BM, bone marrow.
Statistical analysis
All experiments were performed in triplicate by technical replicates. All results were showed as mean ± standard deviation. ANOVA or nonparametric test and Pearson correlation analysis were used in this study. Graphpadprism7 was used for statistical analysis, P values<0.05 were considered statistically significant.
Results
Identification of differentially expressed genes
For the dataset GSE53482, a total of 675 genes (corresponding to 932 transcripts) were thought differentially expressed. Among them, 260 were downregulated and 415 were upregulated.
PPI network construction and sub-network GO&KEGG functional annotation
The PPI network of the differentially expressed genes was constructed and 41 genes were found as hub genes. We finally focused on the sub-network with Itch as hub gene (degree=19) and total 59 nodes (Figure 1). We perform the GO&KEGG functional annotation analyses of the sub-network (Table 2) and observed that the genes engaged in the sub-network were mainly enriched in the PI3K/Akt pathway and several others.
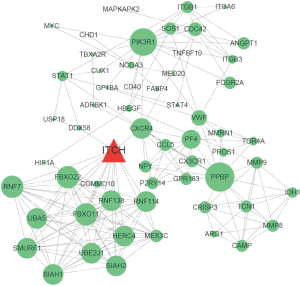
Table 2
| Term | Count | P value | Genes engaged in the terms |
|---|---|---|---|
| GO analysis | |||
| GO:0061630–ubiquitin protein ligase activity | 10 | 8.49E-09 | CDC42, RNF114, RNF7, UBE2J1, RNF138, SIAH1, SIAH2, SMURF1, ITCH, MED20 |
| GO:0004842–ubiquitin-protein transferase activity | 9 | 1.05E-05 | RNF114, RNF7, HERC4, SIAH1, SIAH2, SMURF1, ITCH, FBXO22, FBXO11 |
| GO:0042787–protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 6 | 1.55E-04 | RNF7, HERC4, SIAH1, SIAH2, SMURF1, ITCH |
| GO:0050900–leukocyte migration | 8 | 1.55E-07 | ITGA6, SOS1, MMP9, ANGPT1, ITGB3, PROS1, ITGB1, PIK3R1 |
| GO:0030168–platelet activation | 6 | 3.99E-05 | VWF, PF4, GP1BA, CD40, ITGB3, PIK3R1 |
| KEGG analysis | |||
| hsa04120: Ubiquitin mediated proteolysis | 6 | 9.73E-04 | RNF7, UBE2J1, HERC4, SIAH1, SMURF1, ITCH |
| hsa05200: Pathways in cancer | 10 | 2.71E-04 | CDC42, HIF1A, ITGA6, CXCR4, SOS1, MMP9, STAT1, MYC, ITGB1, PIK3R1 |
| hsa04510: Focal adhesion | 7 | 9.58E-04 | CDC42, VWF, ITGA6, SOS1, ITGB3, ITGB1, PIK3R1 |
| hsa04512: ECM-receptor interaction | 5 | 1.40E-03 | VWF, ITGA6, GP1BA, ITGB3, ITGB1 |
| hsa04151: PI3K-Akt signaling pathway | 8 | 2.88E-03 | VWF, ITGA6, SOS1, ANGPT1, ITGB3, MYC, ITGB1, PIK3R1 |
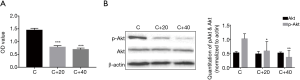
PI3K inhibitor LY294002 could inhibit the proliferation of K562 cells and the activation of Akt
In order to figure out the relationship between the PI3K/Akt pathway and Itch, we first used the PI3K inhibitor LY294002 to treat the K562 cell line. We found that groups treated with LY294002 20 and 40 µmol/L (group C+20, C+40) had less O.D. value when compared with control (group C) by CCK-8 assay. The O.D. value of C, C+20, C+40 was 1.45±0.07, 0.79±0.06, 0.69±0.06 respectively. There was significant difference between group C and the treated groups (P<0.001, Figure 2A) and WB assay found that the activation of Akt decreased in the treated groups (Figure 2B).
LY294002 could induce apoptosis at high concentration and lead to cell cycle arrest of K562 cells
LY294002 could induce apoptosis and cell cycle arrest in tumor cells (19,20). So we used the Flow Cytometry technology to detect about this. The apoptosis percentage of cells of group C, C+20, C+40 (%) was 6.33±0.21, 6.33±0.15, 7.83±0.21 respectively (Figure 3A). There was significant difference between group C and C+40, C+20 and C+40 (P<0.01) but not of the others (P>0.05). The ratio of anti-apoptotic protein bcl-2 and bax had the same change in the treated group though the ratio of cleaved caspase-3 and pro-caspase-3 had no significant difference (P>0.05; Figure 3B). This might illustrate that high concentration of LY294002 could induce apoptosis of K562 cell. The proportion of cells in phage G0/G1, S and G2/M of each group all had significant difference (P<0.01; Figure 3C, Table 3). The proportion of cells in phage G0/G1 of the treated groups increased while the proportion of cells in phage S and G2/M decreased when compared with control and these effects were dose-dependent. The protein p27 suppress the process of cell cycle, restraining cells in G1 phage, inhibiting cells proliferation (21,22). We could find its expression increased in the treated groups but didn’t have significant difference when compared with control (Figure 3D).
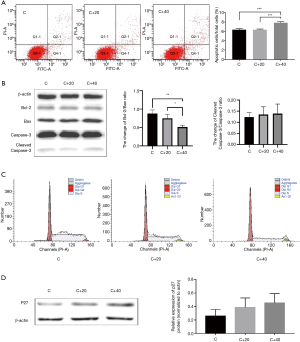
Table 3
| G0/G1 (%) | S (%) | G2/M (%) | |
|---|---|---|---|
| C | 29.5±1.55 | 68.5±1.37 | 1.99±1.76 |
| C+20 | 42.64±1.98*** | 53.49±2.51*** | 3.87±0.53* |
| C+40 | 53.2±2.74**** | 39.31±3.16**** | 7.49±0.89** |
All the results were showed as mean ± standard deviation. *, P<0.05; **, P<0.005; ***, P<0.001; ****, P<0.0001. Group C, the control group.
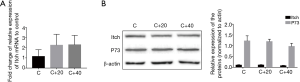
Western blot and Real-time PCR didn’t show significant differences in the expression of Itch & p73 between control and the treated groups
The relative expression level of Itch mRNA of each group (C, C+20, C+40) had no significant difference (P>0.05, Figure 4A) and so was the expression at its protein level. The similar results were found in its substrate p73 (Figure 4B).
The mRNA expression level of Itch increased in patients with fibrosis, high risk or higher WBC count MDS and AML
We examined the mRNA of clinical samples and found that in patients with fibrosis, high risk or higher WBC count MDS and AML, Itch mRNA level significantly increased when compared with the controls (P<0.05, Figure 5A,B,C,D). Through correlative analysis we got that the mRNA level had positive correlation with the WBC count (P<0.001, Figure 5E) (two samples were excluded because the WBC count of them was extremely high than usual and might have a blood test error).
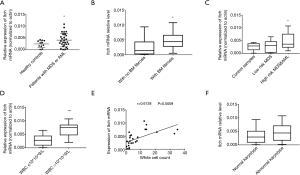
But the mRNA level didn’t show significant difference between the subgroups classified by the karyotype (P>0.05, Figure 5F).
Discussion
In this study, we used the K562 cells which could express both Itch and the PI3K/Akt pathway proteins as a model to investigate whether the pathway regulates the expression of Itch. We found that there was no significant difference in both mRNA and protein level of Itch between groups treated with LY294002 and not. The similar results were found at its substrate p73. Therefore, we figured that the PI3K/Akt pathway might not regulate the expression of Itch in K562 cells and the myeloid tumors. But it probably has other effects on Itch as reported before: The activation of the pathway might not change the expression level of Itch but perform post-transcriptional modification like ubiquitination or phosphorylation on Itch. Panner et al. found that p-Akt could upregulate the ubiquitination level of Itch indirectly and subsequently decrease the interaction between Itch and its substrate (17). Chang et al. reported that p-Akt could phosphorylate the PRR (proline-rich region) domain of Itch at S257 and then Itch would translocate into the nuclear (18) and led to the ubiquitination of H1.2 at K46. Impaired Itch nuclear translocation and H1.2 polyubiquitination sensitized cells to replication stress and limited cell growth and migration.
Chang et al. said that it was JNK (c-Jun N-terminal kinase) but not Akt had mediated the phosphorylation and activation of Itch and finally induced the proteasomal degradation of the protein (23). The similar mechanism was illustrated in pancreatic and breast cancer cells and the process of T cell differentiation (24-26). In future works we will go on studying whether the possible mechanism mentioned above exists in K562 cell line.
Finally, we assayed Itch mRNA level in the clinical samples and found that in patients with high risk MDS, AML, fibrosis or higher WBC count, Itch mRNA level significantly increased when compared with the control groups. The mRNA level of Itch had positive correlation with the WBC count. Therefore, we speculate that Itch may play a role both in the proliferation and the generation of fibrosis in myeloid malignancies with BM fibrosis. This is also consistent with its already reported roles—Itch can get involved in TGF-β pathway and may facilitate the generation of fibrosis. Park et al. found that Itch is an important regulator of Smad7 activity and a positive regulator of TGF-β signaling. It also got involved in the TGF-β-induced epithelial-mesenchymal transition and other biological processes which might help form fibrosis (27). Xing et al. found that callus tissue isolated from Itch knockout mice expressed higher levels of osteoblast associated genes like runx2 which could also engage in the organ fibrosis like kidney and lung through the TGF-β pathway or others (28-30). They also reported that Itch could negatively regulate osteoblast differentiation from mesenchymal progenitor cells (31). Cheng et al. also showed that the extracellular vesicle-carried microRNA-27b derived from mesenchymal stem cells accelerated cutaneous wound healing via E3 ubiquitin ligase Itch (32). These two studies both suggest us Itch may play important roles in mesenchymal cells which have connection with the BM fibrosis. Besides its ability to get involved in the fibrosis formation, Itch could also regulate the tumorgenesis. It can bind p73 and accelerate its degradation, then promote the survival of the tumor cells in the myeloid diseases. The Hippo signaling pathway plays an essential role in the human life. Abnormal regulation of Hippo signaling can be a cause for multiple types of human cancers. It was also reported that the Hippo signaling pathway could be inhibited by Itch which targeted LATS1/2 (large tumor suppressor 1 and 2) for degradation and finally promoted tumorigenicity (33). All these researches increase the possibility of our hypothesis raised above and it as a potential treatment target. Not liking the cases of proteasome, E1 or E2 inhibitors, E3 inhibitors may lead to fewer toxic side effects and to a more suitable targeted therapy through specific targeting of a limited set of substrates theoretically (34,35). So we expect more attention would be focus on this area.
Conclusions
The PI3K/Akt pathway may not get involved in the regulation of the expression of Itch in K562 cells or myeloid tumors and Itch may be a potential target while playing a role both in the proliferation and the generation of fibrosis in myeloid malignancies. In the future, we will do more research on the role of Itch in MDS-F and AML-F and may get more insights of these diseases.
Acknowledgments
We would like to thank Central Lab, Shanghai Sixth People’s Hospital East, Shanghai, China for guidance. We would like to thank Dr. Shiqiao Peng who helped us to polish our manuscript.
Funding: This work was supported by a grant from
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-3115
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-3115
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3115). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients enrolled in this study provided written informed consent to the use of their samples, and this study was approved by the Ethics Committee of the Shanghai Sixth People 's Hospital East, China (No. 2020-089).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Della Porta MG, Malcovati L, Boveri E, et al. Clinical relevant of bone marrow fibrosis and CD34-positive cell clusters in primary myelodysplastic syndromes. J Clin Oncol 2009;27:754-62. [Crossref] [PubMed]
- Fu B, Ok CY, Goswami M, et al. The clinical importance of moderate/severe bone marrow fibrosis in patients with therapy-related myelodysplastic syndromes. Ann Hematol 2013;92:1335-43. [Crossref] [PubMed]
- Fu B, Jaso JM, Sargent RL, et al. Bone marrow fibrosis in patients with primary myelodysplastic syndromes has prognostic value using current therapies and new risk stratification systems. Mod Pathol 2014;27:681-9. [Crossref] [PubMed]
- Tefferi A. Pathogenesis of myelofibrosis with myeloid metaplasia. J Clin Oncol 2005;23:8520-30. [Crossref] [PubMed]
- Kleine-Eggebrecht N, Staufner C, Kathemann S, et al. Mutation in ITCH Gene Can Cause Syndromic Multisystem Autoimmune Disease With Acute Liver Failure. Pediatrics;19;143:e20181554.
- Lorenz S, Cantor AJ, Rape M, et al. Macromolecular juggling by ubiquitylation enzymes. BMC Biol 2013;11:65. [Crossref] [PubMed]
- Fang D, Elly C, Gao B, et al. Dysregulation of T lymphocyte function in itchy mice: A role for Itch in TH2 differentiation. Nat. Immunol 2002;3:281-7. [Crossref] [PubMed]
- Venuprasad K, Zeng M, Baughan SL, et al. Multifaceted role of the ubiquitin ligase Itch in immune regulation. Immunol. Cell Biol 2015;93:452-60. [Crossref] [PubMed]
- Wegierski T, Hill K, Schaefer M, et al. The HECT ubiquitin ligase AIP4 regulates the cell surface expression of select TRP channels. EMBO J 2006;25:5659-69. [Crossref] [PubMed]
- Courbard JR, Fiore F, Adélaïde J, et al. Interaction between two ubiquitin-protein isopeptide ligases of different classes, CBLC and AIP4/ITCH. J. Biol. Chem 2002;277:45267-75. [Crossref] [PubMed]
- Angers A, Ramjaun AR, McPherson PS, et al. The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J.Biol. Chem 2004;279:11471-9. [Crossref] [PubMed]
- Bai Y, Yang C, Hu K, et al. Itch E3 ligase-mediated regulation of TGF-beta signaling by modulating smad2 phosphorylation. Mol. Cell 2004;15:825-31. [Crossref] [PubMed]
- Rossi M, De Laurenzi V, Munarriz E, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J 2005;24:836-48. [Crossref] [PubMed]
- Sampath D, Calin GA, Puduvalli VK, et al. Specific activation of microRNA106b enables the p73 apoptotic response in chronic lymphocytic leukemia by targeting the ubiquitin ligase Itch for degradation. Blood 2009;113:3744-53. [Crossref] [PubMed]
- Fruman DA, Chiu H, Hopkins BD, et al. The PI3K Pathway in Human Disease. Cell 2017;170:605-35. [Crossref] [PubMed]
- Dos Santos C, Récher C, Demur C, et al. The PI3K/Akt/mTOR pathway: a new therapeutic target in the treatment of acute myeloid leukemia. Bull Cancer 2006;93:445-7. [PubMed]
- Panner A, Crane CA, Weng C, et al. Ubiquitin-specific protease 8 links the PTEN-Akt-AIP4 pathway to the control of FLIPS stability and TRAIL sensitivity in glioblastoma multiforme. Cancer Res 2010;70:5046-53. [Crossref] [PubMed]
- Chang L, Shen L, Zhou H, et al. ITCH nuclear translocation and H1.2 polyubiquitination negatively regulate the DNA damage response. Nucleic Acids Res 2019;47:824-42. [Crossref] [PubMed]
- Semba S, Itoh N, Ito M, et al. The in vitro and in vivo effects of 2-(4-morpholinyl)-8-phenyl-chromone (LY294002), a specific inhibitor of phosphatidylinositol 3'-kinase, in human colon cancer cells. Clin Cancer Res 2002;8:1957-63. [PubMed]
- Hu L, Zaloudek C, Mills GB, et al. In vivo and in vitro ovarian carcinoma growth inhibition by a phosphatidylinositol 3-kinase inhibitor (LY294002). Clin Cancer Res 2000;6:880-6. [PubMed]
- Kuo MY, Hsu HY, Kok SH, et al. Prognostic role of p27(Kip1) expression in oral squamous cell carcinoma in Taiwan. Oral Oncol 2002;38:172-8. [Crossref] [PubMed]
- Koguchi K, Nakatsuji Y, Nakayama K, et al. Modulation of astrocyte proliferation by cyclin-dependent kinase inhibitor p27(Kip1). Glia 2002;37:93-104. [Crossref] [PubMed]
- Chang L, Kamata H, Solinas G, et al. The E3 ubiquitin ligase itch couples JNK activation to TNF alpha-induced cell death by inducing c-FLIP(L) turnover. Cell 2006;124:601-13. [Crossref] [PubMed]
- Le Clorennec C, Lazrek Y, Dubreuil O, et al. The anti-HER3 (ErbB3) therapeutic antibody 9F7-F11 induces HER3 ubiquitination and degradation in tumors through JNK1/2- dependent ITCH/AIP4 activation. Oncotarget 2016;7:37013-29. [Crossref] [PubMed]
- Gallagher E, Gao M, Liu YC, et al. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci USA 2006;103:1717-22. [Crossref] [PubMed]
- Venuprasad K, Elly C, Gao M, et al. Convergence of Itch-induced ubiquitination with MEKK1-JNK signaling in Th2 tolerance and airway inflammation. J Clin Invest 2006;116:1117-26. [Crossref] [PubMed]
- Park SH, Jung EH, Kim GY, et al. Itch E3 ubiquitin ligase positively regulates TGF-β signaling to EMT via Smad7 ubiquitination. Mol Cells 2015;38:20-5. [Crossref] [PubMed]
- Liu J, Li X, Zhang H, et al. Ubiquitin E3 ligase Itch negatively regulates osteoblast function by promoting proteasome degradation of osteogenic proteins. Bone Joint Res 2017;6:154-61. [Crossref] [PubMed]
- Kim JI, Jang HS, Jeong JH, et al. Defect in Runx2 gene accelerates ureteral obstruction-induced kidney fibrosis via increased TGF-β signaling pathway. Biochim Biophys Acta 2013;1832:1520-7. [Crossref] [PubMed]
- Mümmler C, Burgy O, Hermann S, et al. Cell-specific expression of runt-related transcription factor 2 contributes to pulmonary fibrosis. FASEB J 2018;32:703-16. [Crossref] [PubMed]
- Zhang H, Xing L. Ubiquitin e3 ligase itch negatively regulates osteoblast differentiation from mesenchymal progenitor cells. Stem Cells 2013;31:1574-83. [Crossref] [PubMed]
- Cheng S, Xi Z, Chen G, et al. Extracellular vesicle-carried microRNA-27b derived from mesenchymal stem cells accelerates cutaneous wound healing via E3 ubiquitin ligase ITCH. J Cell Mol Med 2020;24:11254-71. [Crossref] [PubMed]
- Salah Z, Melino G, Aqeilan RI. Correction: Negative Regulation of the Hippo Pathway by E3 Ubiquitin Ligase ITCH Is Sufficient to Promote Tumorigenicity. Cancer Res;19;79:3007.
- Landré V, Rotblat B, Melino S, et al. Screening for E3-ubiquitin ligase inhibitors: challenges and opportunities. Oncotarget 2014;5:7988-8013. [Crossref] [PubMed]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem 2012;81:167-76. [Crossref] [PubMed]


