
Expression patterns and clinical significances of PBK in lung cancer: an analysis based on Oncomine database
Introduction
Among all malignant tumors, the morbidity and mortality rates of lung cancer (LC) remain some of the highest (1). It is estimated that LC will account for 12% of all global cancers in 2020, and approximately 1% to 4% of cancer deaths are caused by LC (2). LC is comprised of two primary subtypes, namely, small-cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC can be further categorized into three histologic subtypes: squamous-cell carcinoma (SCC), adenocarcinoma (AC) and large-cell lung carcinoma (LCC) (3). Usually, the symptoms of LC are not obvious until the disease enters the advanced stage, and most patients have metastatic or locally advanced diseases at diagnosis (4). These factors are responsible for the dismal prognosis of treatment for individuals with LC.
At present, molecular-targeted drugs have been shown to have broad application prospects in the clinical treatment of lung cancer (5). PDZ-binding kinase (PBK) encodes a serine/threonine protein kinase, which is related to the bispecific mitogen-activated protein kinase (MAPKK) family and participates in many cell processes, such as proliferation, differentiation, transcription regulation and development (6). Our study (7) found that PBK, as a key gene related to the prognosis of LC, is closely related to signal pathways such as cell proliferation, immunity and angiogenesis (8), and has a good clinical research prospect. Therefore, we speculate that PBK is a critical participant in the progression of LC. Presently, few reports have documented the relationship of LC with PBK. Following the rapid development of big data as well as high-throughput sequencing technology cancer-related databases have been being widely used. Herein, we explored the expression mode of PBK in LC along with its potential in prognosis via the Oncomine as well as the Kaplan-Meier Plotter web resources, respectively. We provide a theoretical basis for validating the prospective mechanism of LC development, with a view to discovering new molecular targets for LC, yielding reference for the design of new drugs, and improving the efficacy as well as the quality of life of individuals with LC. Approval was waived by the local ethics committee, as Oncomine database and Kaplan-Meier plotter database are publicly available and de-identified.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-3435).
Methods
Data acquisition and screening
We employed the Oncomine database (https://www.oncomine.org//resource//login) to filter existing data as follows: (I) gene: PBK; (II) cancer type: lung cancer; (III) data type: all; (IV) analysis type: cancer vs. normal analysis; (V) P value <0.001, fold change >2, gene rank = top 10%. Significant difference was set at P<0.05.
Kaplan-Meier Plotter database survival analysis
The survival analysis data of individuals with LC was retrieved through the Kaplan-Meier Plotter database (http://kmplot.com/analysis/): (I) type of cancer: lung cancer; (II) gene: PBK; (III) split patients by auto select best cutoff; (IV) survival: OS. P<0.05 signified a significant difference.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
Survival curve is generated by Kaplan-Meier plots. The results generated in Oncomine showed P values, fold changes and grades. The results of Kaplan-Meier plot showed that HR and P<0.05 were considered to be statistically significant.
Results
Expression of PBK in different tumors
Through the retrieval of the Oncomine web resource, we obtained 291 studies on the expression differences of PBK in different tumor types. PBK expression was remarkably different between the tumor and healthy tissues in 80 studies; of those 80, PBKs were upregulated in 63 studies, while they were downregulated in 17 studies. As indicated in Figure 1, downregulated PBK was documented in breast cancer (n=1), leukemia (n=5), leukemia (n=1) and others (n=6). Upregulated PBK was documented in head and neck cancer (n=3), brain and CNS cancer (n=6), breast cancer (n=12), cervical cancer (n=2), colorectal cancer (n=2), esophageal cancer (n=2), gastric cancer (n=5), liver cancer (n=3), lung cancer (n=8), lymphoma (n=4), sarcoma (n=9), ovarian cancer (n=3), pancreatic cancer (n=1), prostate cancer (n=3), bladder cancer (n=2), and others (n=3) (Figure 1).
Expression pattern of PBK in lung cancer tissue
There were seven studies on the analysis of expression differences between PBK in lung cancer tissues and healthy tissues including 774 samples. These seven studies were performed by Hou (9), Garber (10), Su (11), Landi (12), Selamat (13), Wachi (14) and Okayama (15). Meta-analysis results showed an upward adjustment in PBK expression in LC tissues relative to healthy tissues (median rank =752.0, P=5.53E−11) (Figure 2).
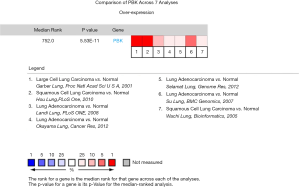
Difference expression of PBK in microarrays of lung cancer
In the seven microarray data of Hou (9), Garber (10), Su (11), Landi (12), Selamat (13), Wachi (14) and Okayama (15), the PBK level in the LC tissue is remarkably higher than it is in healthy tissue (P<.05) (Figure 3).
Potential value of PBK in the prognosis of lung cancer
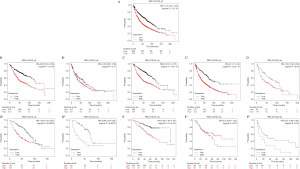
Survival analysis on the basis of the Kaplan-Meier Plotter was conducted to determine the prognostic potential of PBK in LC. Differential PBK expression levels in individuals with LC were significantly correlated with prognosis. The data demonstrate that the expression level of PBK is inversely linked to the total survival period of individuals with LC. Patients with LC with high PBK expression have a poor survival period (P<0.01). The subgroup analysis was as follows: (I) In terms of histological classification, PBK was linked to the survival period of AC patients (P<0.01), and there was no correlation with SCC (P=0.12). (II) From the perspective of the patient's sex, the survival period of the female and male low-expressed group was remarkably higher in contrast with that of the high-expression group, with P values 1.4e-10 and 3.3e-12, respectively (3). Subgroup analysis by differentiated grades showed that the survival period of the PBK low-expression group was remarkably higher in contrast with that of the high-expression group in grade I, II and III LC with P values 0.04, 0.00023 and 0.00073, respectively (4). In terms of clinical stage, the survival time of PBK in the low-expression group was significantly higher than that of the high-expression group in stage 1 and stage 3 lung cancer, and P-values were 2.1e-05 min and 0.03, respectively. There was no remarkable difference in survival between low- and high-expressed groups of stage 2 lung cancer (P=0.23). There is still a lack of data on the difference in survival between PBK low- and high-expression groups in stage 4 LC (Figure 4).
Discussion
CancerSEA (16), the first dedicated database that aims to comprehensively explore distinct functional states of cancer cells at the single-cell level. CancerSEA shows that the functional status of PBK is mainly concentrated in invasion, proliferation, EMT, cell cycle, DNA damage, DNA repair (P≤0.001). These functions are very important in the occurrence and development of tumors. It has been found that PBK is rarely expressed in healthy tissues except in fetuses and germ cells but is highly transient in numerous cancers (17). The high expression of PBK (also known as MAPKK-like protein kinase) is related to the invasive phenotype of multiple cancers (18,19). These attribute make PBK a prospective molecular target for cancer diagnosis as well as targeted treatment (20).
Currently, PBK is attracting researcher attention given the increasing amount of research evidence demonstrating that it has a pivotal role in carcinogenesis as well as metastasis. Xu et al. (21) have shown that in many tumors, the expression of PBK is often upgraded, which is significantly related to the patient's low survival rate. Dou et al. (22) found that atorvastatin or vanilla transferase I inhibitor GGTI-298 inhibits breast cancer cell proliferation by inactivated YAP signals and lowering PBK. Ohashi et al. (23) found that PBK serves as a carcinogen by overexpression in esophageal squamous cell carcinoma, which represses cell proliferation and infiltration when PBK/TOPK is downregulated. Kwon et al. (24) verified by real-time fluorescence quantitative reverse transcription-polymerase chain reaction evaluation that among the differentially expressed genes studied, the upregulated expression of proteasome β 8 as well as PBK can enhance the migration along with the infiltration of gastric cancer cells and can act as effective biosignatures for diagnosing patients with dismal prognosis.
Given the small number of single research samples and a certain degree of systemic bias, the Oncomine web resource emerged. The Oncomine web resource is presently the world's most extensive oncogene chip web resource as well as integrated data mining platform. The Oncomine web resource integrates RNA-seq along with the DNA-seq data from the Cancer Genome Atlas (TCGA) as well as the Gene Expression Omnibus (GEO) and published literature to explore differential expression in cancer and healthy tissues (8). The database not only helps researchers to carry out data mining but also has the function of secondary analysis, which has made a great contribution to the search and functional analysis of tumor differential genes.
This study employed independent data sets from the Oncomine database and Kaplan-Meier plotter databases to detect the expression levels of PBK and prognostic landscape in lung cancer. Here, we used Oncomine web resource to explore the differential expression of PBK in different tumor tissues. In 80 studies documenting the marked difference in PBK expression, 63 studies reported upregulated PBK in various kinds of tumor tissues, including bladder cancer, head and neck cancer, brain and CNS cancer, colorectal cancer, gastric cancer, liver cancer, lung cancer, lymphoma, ovarian cancer, Esophageal cancer, cervical cancer, pancreatic cancer, breast cancer, prostate cancer, and sarcoma. In addition, upregulation of PBK was found in 774 individuals with LC, constituting AC, SCC and LCC. The prognostic value of PBK in LC was analyzed through the Kaplan-Meier Plotter web platform. Our study found that the expression level of PBK was negatively linked to the overall survival of LC. The increased expression of PBK indicates a poor prognosis. Subgroup assessment showed that the prognostic significance of PBK was more significant in patients with lung adenocarcinoma. In regards to gender, grade I-III differentiation, and clinical stages 1 and 3, the survival advantage of the PBK high-expression group was remarkably lower than that of the low-expression group. Nevertheless, there was no significant difference in the expression of PBK in stage 2 lung cancer. Because of the lack of expression data in stage 4 lung cancer, it is necessary to expand further the sample size for research and analysis.
In summary, this study explored the expression trend as well as prognostic ability of PBK in patients with LC via the Oncomine as well as Kaplan-Meier Plotter web resources. We found that PBK expression is upregulated in individuals with LC, while the high expression of PBK is disadvantageous to the OS of LC. Our conclusions may provide a new way to explore the mechanism of LC, to develop biomarkers for diagnosis or prognosis and to target drugs. It should also be emphasized that there are still some limitations in this study. First, the heterogeneity between studies cannot be ignored. Second, in some studies, a small sample size may have led to deviation. Third, since all data are only from the Oncomine database, the conclusions may be limited. We will, moreover, validate the role of PBK in LC through cytological as well as animal model studies in the later stage and analyze the clinical and pathological correlation between PBK expression and LC.
Acknowledgments
We thank AJE (https://www.aje.cn) for its linguistic assistance during the preparation of this manuscript.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-3435
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3435). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Doroshow D, Herbst R. Treatment of Advanced Non-Small Cell Lung Cancer in 2018. JAMA Oncol 2018;4:569-70. [Crossref] [PubMed]
- Barrón-Barrón F, Guzmán-De Alba E, Alatorre-Alexander J, et al. National Clinical Practice Guidelines for the management of non-small cell lung cancer in early, locally advanced and metastatic stages. Extended version. Salud Publica Mex 2019;61:359-414. [PubMed]
- Dong A, Zhao Y, Li Z, et al. PD-L1 vs. Tumor mutation burden, which is the better immunotherapy biomarker in advanced non-small cell lung cancer? J Gene Med 2020;e3294. [Crossref]
- Dougherty JD, Garcia ADR, Nakano I, et al. PBK/TOPK, a proliferating neural progenitor-specific mitogen-activated protein kinase kinase. J Neurosci 2005;25:10773-85. [Crossref] [PubMed]
- Li J, Hu S, Zhou T, et al. Comprehensive analysis of the competing endogenous RNA network in lung adenocarcinoma: Base on TCGA database. Journal of Modern Oncology 2020;28:4220-6.
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 2010;5:e10312. [Crossref] [PubMed]
- Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 2001;98:13784-9. [Crossref] [PubMed]
- Su LJ, Chang CW, Wu YC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 2007;8:140. [Crossref] [PubMed]
- Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One 2008;3:e1651. [Crossref] [PubMed]
- Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012;22:1197-211. [Crossref] [PubMed]
- Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics 2005;21:4205-8. [Crossref] [PubMed]
- Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 2012;72:100-11. [Crossref] [PubMed]
- Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res 2019;47:D900-8. [Crossref] [PubMed]
- Herbert KJ, Ashton TM, Prevo R, et al. T-LAK cell-originated protein kinase (TOPK): an emerging target for cancer-specific therapeutics. Cell death & disease 2018;9:1089. [Crossref] [PubMed]
- Simons-Evelyn M, Bailey-Dell K, Toretsky J, et al. PBK/TOPK is a novel mitotic kinase which is upregulated in Burkitt's lymphoma and other highly proliferative malignant cells. Blood cells, molecules & diseases 2001;27:825-9. [Crossref] [PubMed]
- Zhao S, Dai J, Zhao W, et al. PDZ-binding kinase participates in spermatogenesis. The international journal of biochemistry & cell biology 2001;33:631-6. [Crossref] [PubMed]
- Ma H, Li Y, Wang X, et al. PBK, targeted by EVI1, promotes metastasis and confers cisplatin resistance through inducing autophagy in high-grade serous ovarian carcinoma. Cell Death Dis 2019;10:166. [Crossref] [PubMed]
- Xu M, Xu S. PBK/TOPK overexpression and survival in solid tumors: A PRISMA-compliant meta-analysis. Medicine (Baltimore) 2019;98:e14766. [Crossref] [PubMed]
- Dou X, Wei J, Sun A, et al. PBK/TOPK mediates geranylgeranylation signaling for breast cancer cell proliferation. Cancer Cell Int 2015;15:27. [Crossref] [PubMed]
- Ohashi T, Komatsu S, Ichikawa D, et al. Overexpression of PBK/TOPK Contributes to Tumor Development and Poor Outcome of Esophageal Squamous Cell Carcinoma. Anticancer Res 2016;36:6457-66. [Crossref] [PubMed]
- Kwon CH, Park HJ, Choi YR, et al. PSMB8 and PBK as potential gastric cancer subtype-specific biomarkers associated with prognosis. Oncotarget 2016;7:21454-68. [Crossref] [PubMed]




