
Up-regulation of SOCS4 promotes cell proliferation and migration in esophageal squamous cell carcinoma
Introduction
Esophageal cancer (EC) is the sixth most common malignancy worldwide (1), with esophageal squamous cell carcinoma (ESCC) accounting for ~90% of ECs (1). Despite improvements in ESCC treatments in recent years, the cure rate of ESCC is still low (2,3). Most ESCC is diagnosed at advanced stage, which makes treatments more difficult and less effective (4). Unfortunately, the mechanism underlying ESCC metastasis remains unknown. Therefore, studies are needed to identify the proteins that play critical roles in regulating the metastasis of ESCC, which is essential for improving early detection and the discovery of novel therapeutic targets.
Suppressors of cytokine signaling (SOCS) family proteins are negative modulators of cytokine and growth factor signaling. The SOCS family consists of eight members, including SOCS1–SOCS7 and cytokine-induced signal transducer and activator of transcription (STAT) inhibitor (CIS) (5,6). The loss or down-regulation of SOCS proteins has been shown to lead to many diseases, including cancer. In the SOCS family, SOCS1 and SOCS3 have been reported in different cancers, including gastric cancer (7), laryngeal carcinoma (8), hepatocellular carcinoma (9), melanoma (10), and breast cancer (11-13). Recently, the down-regulation of SOCS4 was shown to serve critical and multifaceted roles in the progression of numerous cancers, including hepatocellular carcinoma (14), thyroid cancer (15), breast cancer, and lung adenocarcinoma (16). These findings indicate that SOCS4 may be an attractive therapeutic target in cancer treatment. While, the expression and the roles of SOCS4 in ESCC remain elusive.
In the current report, the expression of SOCS4 in ESCC was detected, and the relationship between SOCS4 expression and the clinicopathological features of ESCC was investigated. Moreover, the roles of SOCS4 on the proliferation and migration of ESCC cells were also investigated.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-21-700).
Methods
Bioinformatic analysis
The datasets in The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) were used to examine the expression pattern of SOCS4 in ESCC. Briefly, we used GEPIA (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/index.html), which is a web-based tool, to perform the differential gene analysis (tumor: n=182; normal: n=286) in TCGA.
Patients and tissue samples
A total of 103 ESCC tissues and non-carcinoma tissues were obtained from the Affiliated Nanjing Hospital of Nanjing Medical University. All patients underwent surgery between July 2006 and December 2008, and did not receive any anti-tumor therapy prior to surgery. The overall survival (OS) of patients denotes the months from diagnosis until the date of death, last known to be alive, or the study closing date. The clinicopathological parameters of the patients are shown in Table 1. This study was approved by the Ethics Committee of People’s Hospital of Xuyi (No. XYLL-2018006). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients.
Table 1
| Variables | Patient (n=103) |
|---|---|
| Age (y ± SD) | 65.2±9.3 |
| Sex (female: male) | 28:75 |
| Tumor size | |
| <5 cm | 59 |
| ≥5 cm | 44 |
| Pathological grade | |
| Poor | 8 |
| Moderately | 67 |
| Well | 28 |
| Lymph node (LN) metastasis | |
| Yes | 38 |
| No | 65 |
ESCC, esophageal squamous cell carcinoma.
Immunohistochemical (IHC) staining
For immunohistochemistry, paraffin sections were deparaffinised, and then rehydrated with graded ethyl alcohol. Hydrogen peroxide (3%) was used to block endogenous peroxidase activity. A sodium citrate buffer was used to retrieve the antigens. A rabbit polyclonal antibody targeting SOCS4 (cat. No. PA5-21599; 1:1,000 dilution; Thermo Fisher Scientific™ Invitrogen, USA) was incubated at 4 °C overnight, and a horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG) (cat. No. ab6721, 1:1,000 dilution, Abcam) was incubated at room temperature for 1 h. The slides were then developed with diaminobenzidine (DAB) (Beyotime Institute of Biotechnology, Haimen, China) for 5 min at room temperature. Images were captured under a light microscope (Olympus Corporation, Tokyo, Japan).
The stains were evaluated separately by two pathologists under double-blinded conditions and scored by staining intensity and positive staining. Staining intensity was graded as negative (0 point), weak (1 point), moderate (2 points), or strong (3 points). Positive staining was quantified and classified into five categories scored 0 to 4: <10% positive cells as 0; 10–25% as 1; 26–50% as 2; 51–75% as 3, and ≥76% as 4. The final score was calculated as the staining intensity score multiplied by the positive staining score. For categorical analyses, final scores of expression levels greater than 4 (final score >4) were regarded as high.
Cell culture
Eca-109 (human ESCC cell line) was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and was cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific™, USA) supplemented with 10% foetal bovine serum (YEASEN, Shanghai, China) and 100 U/mL penicillin/streptomycin (Gibco, Thermo Fisher Scientific™, USA) in 5% CO2 incubator at 37 °C.
siRNA transfection
The siRNA of SOCS4 was designed by the Shanghai China, GenePharma Co., Ltd., and the siRNA sequences are listed in Table 2. The siRNA was transfected into Eca109 cells using Lipofectamine RNA iMAX (Thermo Fisher Scientific™, USA) according to the manufacturer’s instructions. The cells were harvested 48–72 h after transfection and analyzed by quantitative real-time polymerase chain reaction (RT-qPCR) and western blotting.
Table 2
| Tissue | SOCS4 expression level | P value | |
|---|---|---|---|
| High | Low | ||
| ESCC tissue | 56 | 47 | <0.001 |
| Non-carcinoma tissues | 17 | 49 | |
ESCC, esophageal squamous cell carcinoma.
Cell proliferation assay
After siRNA transfection for 48 h, the cells were harvested and seeded in 6 cm dishes (1×104 cells/dish). The cell number was then counted every 24 h for a total of 5 days. A cell growth curve, expressed as the fold change in cell growth, was created using GraphPad Prism 5 (GraphPad software, Inc., USA).
Cell counting kit-8 (CCK-8) assay
A CCK-8 assay was used to measure cell viability. After siRNA transfection for 48 h, the cells were seeded in 96-well plates triplicatedly (1.0×104 cells/mL). After 24 h, cell viability was measured using CCK-8 (Nanjing KeyGen Biotech Co., Ltd. China) according to the manufacturer’s instructions. The absorbance was measured at 450 nm using a microplate reader (Multiskan FC, Thermo, USA), and the data was recorded every 24 h for 5 consecutive days.
Cell cycle analysis
After siRNA transfection, the cells were cultured in a serum-free medium for 24 h to synchronize at the G0/G1 phase, and cells were centrifugated (500 g ×5 min). After washing with phosphate buffered saline (PBS) three times, the cells were fixed with 70% ethanol at 4 °C overnight. Subsequently, the cells were washed with ice-cold PBS twice, and treated with RNA enzyme A (RNase A) (20 µg/mL of final concentration) and propidium iodide (10 µg/mL of final concentration) in the dark for 1 h at room temperature. The cell populations at the G0/G1, S, and G2/M phases were measured by flow cytometry (Navios, Thermo Fisher Scientific™, USA), and quantified using Kaluza Analysis software (Thermo Fisher Scientific™, USA). The assay was conducted triplicatedly.
Wound-healing assay
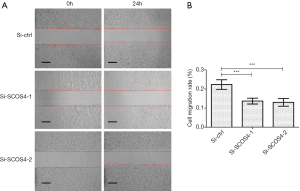
Cell migration ability was examined as previously reported (17). Briefly, the cells were seeded in 6-well plates until they reached confluence, and were then scratched using a 200-µL pipette tip. The gap was measured at 0 h (w1) and at 24 h (w2), respectively, and the relative migration rate was calculated as (w1−w2)/w1 ×100%. The wounds were photographed using a light microscope at a magnification of 100× (CKX53, OLYMPUS, Japan). The assay was conducted triplicatedly.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using the Ultrapure RNA Kit (CWBIO, Beijing, China) according to the manufacturer’s instructions. Next, complementary DNA (cDNA) was synthesized using the 5× All-In-One Master Mix with AccuRT genomic DNA Removal Kit (abm®, Canada; catalog G492). Subsequently, quantitative PCR was performed using BrightGreen 2× qPCR Master Mix-ROX (abm®, Canada; catalog Master Mix-R) with the ABI StepOne Plus (Thermo Fisher ScientificTM, USA). The reaction conditions were as follows: 95 °C 10 min, 95 °C 15 s, and 60 °C 1 min for 40 cycles. The relative quantitative SOCS4 expression value was normalized to that of glyceraldehydes-3-phosphate dehydrogenase (GAPDH) and calculated using the 2-∆∆CT method. Each sample was examined in triplicate. The primers were designed as follows: SOCS4 forward, 5'-CATGGCAGAAAATAATGAAAATAT-3' and reverse, 5'-GTTTAAGAGATCGGCCTAAAAAT-3'; and GAPDH forward, 5'-ACCACAGTCCATGCCATCAC-3' and reverse, 5'-TCCACCACCCTGTTGCTGTA-3'.
Western blot
Cells were lysed in a radio immunoprecipitation assay (RIPA) buffer (Beyotime, China) supplemented with Complete Protease Inhibitor Cocktail (Roche, Germany) plus 1 mM phenylmethanesulfonyl fluoride (PMSF). For tissues, total protein was extracted from ESCC tissues and matched non-carcinoma tissues in 2% sodium dodecyl sulfate (SDS) in Nonidet P-40 (NP-40) lysate buffer, and then boiled at 98 °C for 10 min. After centrifugation (18,000 g ×15 min) at 4 °C, the supernatant was collected for use. Protein concentrations were measured using a BCA Kit (Beyotime, China). Next, 30 µg protein was electrophoresed through a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto polyvinylidene fluoride (PVDF)membranes (Millipore, USA). After blocking with 5% milk, the membranes were immunoprobed with primary antibody at 4 °C overnight. After washing, the membranes were incubated with HRP-conjugated secondary antibody (Proteintech, USA) at room temperature for 1 h. After washing, the membranes were developed using an enhanced ECL kit (Beyotime, China), and the image was detected using a Clinx imaging system (Clinx Science Instruments, Co., Ltd., China). The primary antibodies were as follows: anti-SOCS4 antibody (1:1,000; Thermo Fisher Scientific™ Invitrogen, USA; catalog PA5-21599), anti-ICAM1 antibody (1:1,000; Cell Signaling Technology, USA; catalog #4915), anti-VCAM1 antibody (Cell Signaling Technology, USA; catalog #39236), anti-uPA antibody (1:1,000; Cell Signaling Technology, USA; catalog #67589), anti-TNF-α antibody (1:2,000; Cell Signaling Technology, USA; catalog #6945), anti-NF-κB p65 antibody (1:2,000; Cell Signaling Technology, USA; catalog #8242), anti-phospho-NF-κB p65 antibody (1:2,000; Cell Signaling Technology, USA; catalog #3033), and anti-β-actin antibody (1:3,000; Sigma-Aldrich, Japan; catalog A1978).
Statistical analysis
The data were analysed using SPSS 25.0 (SPSS, Inc., Chicago, IL, USA). All data were recorded as the mean ± SD or n/%. The differences in the expression of SOCS4 between tumour tissues and non-carcinoma tissues were analysed using Chi-square analysis. The correlations between SOCS4 expression and the clinicopathological parameters of ESCC were analysed using Pearson’s chi-squared test. Survival curves were generated using the Kaplan-Meier method and analyzed by log-rank test. The Student’s t-test was used to determine the differences between two groups. One-way analysis of variance (ANOVA) was used to analyze the differences between multiple groups. All experiments were performed at least three times, and P<0.05 was considered statistically significant.
Results
Upregulatedexpression of SOCS4 in human ESCC tissues
First, SOCS4 expression was analyzed in human ESCC tissues. SOCS4 mRNA was significantly upregulated in ESCC tissues compared to adjacent normal tissues (Figure 1A). A total of 5 independent ESCC tissues were then analyzed, and the results showed that the expression levels of the SOCS4 protein in ESCC tissues was significantly higher than that in adjacent normal tissues (Figure 1B).
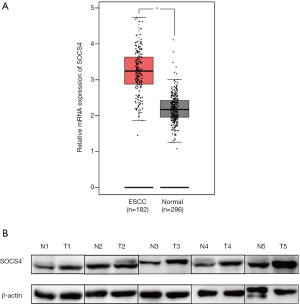
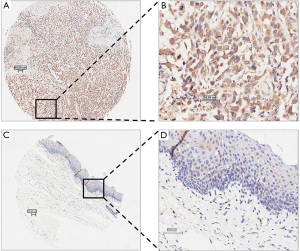
Next, immunohistochemistry was used to further measure the expression of SOCS4 in 103 ESCC tissues. In the ESCC tissues, SOCS4 was mostly expressed in the cytoplasm (Figure 2). Moreover, in ESCC tissues, the positive expression of SOCS4 was 54.4% (56/103), and was only 25.7% (17/66) in the adjacent normal tissues (Table 2).
Correlations between SOCS4 expression and the clinicopathological features of ESCC
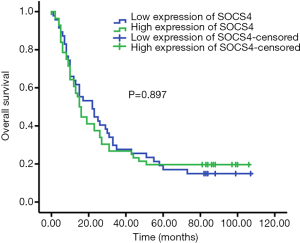
The correlations between SOCS4 expression and the clinicopathological features of ESCC were analyzed. We found that SOCS4 expression was not associated withage, gender, or pathological grade of ESCC. The high-expression of SOCS4 was positively correlated with tumor size and lymph node metastasis (Table 3). Kaplan-Meier survival analysis revealed that ESCC patients with a high SOCS4 expression had a similar OS compared to those with a low SOCS4 expression (Figure 3).
Table 3
| Characteristics | SOCS4 expression level | P value | |
|---|---|---|---|
| Low (%) | High (%) | ||
| All patients | 47 (45.6) | 56 (54.4) | |
| Age (years) | |||
| <65 | 24 (46.2) | 28 (53.8) | 0.915 |
| ≥65 | 23 (45.1) | 28 (54.9) | |
| Gender | |||
| Male | 35 (46.7) | 40 (53.3) | 0.733 |
| Female | 12 (42.9) | 16 (57.1) | |
| Tumor size | |||
| <5 cm | 34 (57.6) | 25 (42.4) | 0.016 |
| ≥5 cm | 13 (28.3) | 31 (71.7) | |
| Pathological grade | |||
| Poor | 4 (50.0) | 4 (50.0) | 0.695 |
| Moderately | 31 (46.3) | 36 (53.7) | |
| Well | 12 (42.9) | 16 (57.1) | |
| Lymph node metastasis | |||
| Yes | 7 (18.4) | 31 (81.6) | 0.006 |
| No | 40 (61.5) | 25 (38.5) | |
SOCS4, suppressors of cytokine signalling family member 4; ESCC, esophageal squamous cell carcinoma.
Knockdown of SOCS4 inhibits the proliferation of ESCC cells in vitro
In order to determine whether SOCS4 plays a vital role in ESCC cell behaviors, SOCS4 was knowdowned by siRNA in Eca109 cells. The expression of SOCS4 mRNA (Figure 4A) and protein (Figure 4B) were successfully decreased in Eca109 cells compared with the control siRNA-transfected cells. Our results showed that SOCS4 knockdown reduced the growth of Eca109 cells when assessed daily over a 5-day period (Figure 4C), and also decreased cell viability in the CCK-8 assay (Figure 4D).
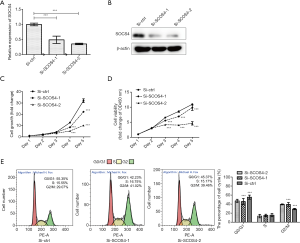
Next, the cell cycle was detected by flow cytometry. SOCS4 knockdown resulted ina decrease in the percentage of G0/G1 phase cells from 55.38% to 42.23% (Si-SOCS4-1) or 45.37% (Si-SOCS4-2), and an increase in the percentage G2/M phase cells from 29.07% to 41.02% (Si-SOCS4-1) or 39.46% (Si-SOCS4-2) (Figure 4E).
Knockdown of SOCS4 inhibits the migration of ESCC cells in vitro
A wound healing assay was performed. The results showed that SOCS4 knockdown in Eca109 cells significantly inhibited cell migration compared with control cells (Figure 5A,B).
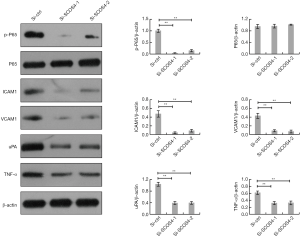
Knockdown of SOCS4 inhibits NF-κB signaling in ESCC cells
It has been well documented that NF-κB signaling is critical for ESCC cell proliferation and migration. The expression level of NF-κB p65 was also quantified by western blotting as shown in Figure 6, the phosphorylated levels of p65 were down-regulated by SOCS4 knockdown, however the total p65 level was unaffected. Moreover, several NF-κB-regulated metastasis-related genes, including intercellular adhesion molecule 1 (ICAM1), vascular cell adhesion molecule 1 (VCAM1), urokinase-type plasminogen activator (uPA), and tumor necrosis factor (TNF-α), were markedly decreased in SOCS4-knockdown ESCC cells.
Discussion
SOCS proteins play important roles in the progression of various tumors (18-21). SOCS4, a major member of the SOCS family, negatively regulates cell signal transduction. Cumulative studies have demonstrated that SOCS4is critical for the progression of multiple types of cancers, including breast cancer (13,22), lung adenocarcinoma (16,23), endometrial carcinoma (24), gastric cancer (25,26), and hepatic carcinoma (27). However, the expression and actual biological functions of SOCS4 in ESCC remain elusive. The current study investigated the expression and functions of SOCS4 in ESCC, which may provide potential implications for ESCC therapy.
In this study, the clinical significance of SOCS4 in ESCC was investigated for the first time. SOCS4 was found to be up-regulated in human ESCC tissues, which was associated with tumor size and lymph node metastasis. These findings indicated that SOCS4 might play a pro-tumor role in ESCC.
Previous studies have reported that SOCS4 is involved in several important processes, including cell migration, proliferation, invasion, and the stemness maintenance of cancer stem cells (14-16). In the current study, we found that the knockdown of SOCS4 could reduce the migration capacity of Eca109 cells. Furthermore, the suppression of SOCS4 reduced the proliferation of Eca109 cells along with cell accumulation in the G2/M phase. These results indicated that SOCS4 promoted cell proliferation and migration in ESCC cells.
The above findings were different from the previous findings on other cancer cells, which showed that SOCS4 is down-regulated in gastric cancer, hepatic carcinoma, breast cancer, and lung adenocarcinoma, and acted as a tumor suppressor in these cancers, inhibiting the proliferation, migration, and invasion of cancer cells (14-16). This discrepancy between our results and these previous findings suggests that the effects of SOCS4 on the malignant behaviors of cancer cells may be cell-type dependent on the heterogeneity of SOCS4, or its interaction with distinct substrates or binding partners.
In addition, we further explored the underlying mechanism. We found that SOCS4 silencing markedly inhibited phosphorylated levels of NF-κB p65 as well as NF-κB-regulated metastasis-related genes, including ICAM1, VCAM1, uPA, and TNFα, suggesting that NF-κB signaling was regulated by SOCS4 in ESCC cells. NF-κB signaling is critical for proliferation and migration in numerous cancer cell types, including ESCC cells (28-30). Therefore, it is reasonable to speculate that the decreased proliferation and migration observed in SOCS4-knockdown Eca109 cells is due to the inactivation of NF-κB signaling. A previous study showed that NF-κB activity was negatively regulated by SOCS4 in IPEC-J2 cells (31), which was contrary to our results. This suggested that the effect of SOCS4 on NF-κB activity depends on cell type. In future research, it will be important to investigate the mechanism through which SOCS4 regulates NF-κB activity in our system.
Previous studies have also demonstrated that SOCS4 inactivated the phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) and Janus kinase/signal transducer and activator of transcription 3 (JAK/STAT3) pathways, and therefore served as a tumor suppressor in several types of cancer cells (14-16). The PI3K/AKT pathway is activated in ESCC and impacts on various biological behaviors such as migration, invasion, proliferation, and apoptosis (32-35). The activation of the JAK/STAT3 pathway plays a critical role in the migration, invasion, and proliferation of ESCC (36-40). Based on these findings, we hypothesized that SOCS4 might promote the proliferation and migration of ESCC cells in a PI3K/AKT and JAK/STAT3 pathways-independent way, which needs to be investigated in the near future.
However, there are some limitations in the current study. First, more than one cell line should be used to investigate the role of SOCS4 on ESCC. Then, further study is needed to investigate the effect of SOCS4 on ESCC cell proliferation and migration in vivo, as well as its underlying mechanisms. Nest, it will also be important to investigate the role and underlying mechanisms of SOCS4 on other aspects of ESCC progression such as apoptosis. Finally, the expression pattern, functions, and molecular mechanism of SOCS4 in other types of cancer are also needed further investigated.
Conclusions
The current study revealed, for the first time, that the up-regulation of SOCS4 in ESCC was associated with tumor size and lymph node metastasis. SOCS4 knockdown in vitro inhibited the proliferation and migration of ESCC cells and NF-κB signaling, indicating that SOCS4 may be a novel potential therapeutic target for ESCC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-21-700
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-21-700
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-21-700). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of People’s Hospital of Xuyi (No. XYLL-2018006). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Abnet CC, Arnold M, Wei WQ. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018;154:360-73. [Crossref] [PubMed]
- González-Plaza JJ, Hulak N, Garcia-Fuentes E, et al. Oesophageal squamous cell carcinoma (ESCC): Advances through omics technologies, towards ESCC salivaomics. Drug Discov Ther 2015;9:247-57. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Ca Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Sun D, Chen C, Hu W, et al. Low expression level of ASK1-interacting protein-1 correlated with tumor angiogenesis and poor survival in patients with esophageal squamous cell cancer. Onco Targets Ther 2018;11:7699-707. [Crossref] [PubMed]
- Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol 2009;30:592-602. [Crossref] [PubMed]
- Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol 2003;4:1169-76. [Crossref] [PubMed]
- Wang L, Zhuang L, Rong H, et al. MicroRNA-101 inhibits proliferation of pulmonary microvascular endothelial cells in a rat model of hepatopulmonary syndrome by targeting the JAK2/STAT3 signaling pathway. Mol Med Rep 2015;12:8261-7. [Crossref] [PubMed]
- Starska K, Forma E, Lewy-Trenda I, et al. The expression of SOCS1 and TLR4-NFkappaB pathway molecules in neoplastic cells as potential biomarker for the aggressive tumor phenotype in laryngeal carcinoma. Folia Histochem Cytobiol 2009;47:401-10. [PubMed]
- Wei L, Huang Y, Zhao R, et al. Detection of promoter methylation status of suppressor of cytokine signaling 3 (SOCS3) in tissue and plasma from Chinese patients with different hepatic diseases. Clin Exp Med 2018;18:79-87. [Crossref] [PubMed]
- Li Z, Metze D, Nashan D, et al. Expression of SOCS-1, suppressor of cytokine signalling-1, in human melanoma. J Invest Dermatol 2004;123:737-45. [Crossref] [PubMed]
- Lv Y, Song G, Li P. Correlation of SOCS-1 gene with onset and prognosis of breast cancer. Oncol Lett 2018;16:383-7. [Crossref] [PubMed]
- Nakagawa T, Iida S, Osanai T, et al. Decreased expression of SOCS-3 mRNA in breast cancer with lymph node metastasis. Oncol Rep 2008;19:33-9. [Crossref] [PubMed]
- Sasi W, Jiang WG, Sharma A, et al. Higher expression levels of SOCS 1,3,4,7 are associated with earlier tumour stage and better clinical outcome in human breast cancer. BMC Cancer 2010;10:178. [Crossref] [PubMed]
- Jiang C, Long J, Liu B, et al. miR-500a-3p promotes cancer stem cells properties via STAT3 pathway in human hepatocellular carcinoma. J Exp Clin Cancer Res 2017;36:99. [Crossref] [PubMed]
- Mei Z, Chen S, Chen C, et al. Interleukin-23 Facilitates Thyroid Cancer Cell Migration and Invasion by Inhibiting SOCS4 Expression via MicroRNA-25. PLoS One 2015;10:e0139456. [Crossref] [PubMed]
- Xiao X, Yang D, Gong X, et al. miR-1290 promotes lung adenocarcinoma cell proliferation and invasion by targeting SOCS4. Oncotarget 2018;9:11977-88. [Crossref] [PubMed]
- Gu Z, Li Y, Yang X, et al. Overexpression of CLC-3 is regulated by XRCC5 and is a poor prognostic biomarker for gastric cancer. J Hematol Oncol 2018;11:115. [Crossref] [PubMed]
- Inagaki-Ohara K, Kondo T, Ito M, et al. SOCS, inflammation, and cancer. JAKSTAT 2013;2:e24053. [Crossref] [PubMed]
- Zhang J, Li H, Yu JP, et al. Role of SOCS1 in tumor progression and therapeutic application. Int J Cancer 2012;130:1971-80. [Crossref] [PubMed]
- Yin Y, Liu W, Dai Y. SOCS3 and its role in associated diseases. Hum Immunol 2015;76:775-80. [Crossref] [PubMed]
- Letellier E, Haan S. SOCS2: physiological and pathological functions. Front Biosci (Elite Ed) 2016;8:189-204. [Crossref] [PubMed]
- Choi J, Song N, Han S, et al. The Associations between Immunity-Related Genes and Breast Cancer Prognosis in Korean Women. PLoS One 2014;9:e103593. [Crossref] [PubMed]
- Ma J, Mannoor K, Gao L, et al. Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol Oncol 2014;8:1208-19. [Crossref] [PubMed]
- Wu X, Cai D, Zhang F, et al. Long noncoding RNA TUSC7 inhibits cell proliferation, migration and invasion by regulating SOCS4 (SOCS5) expression through targeting miR-616 in endometrial carcinoma. Life Sci 2019;231:116549. [Crossref] [PubMed]
- Kobayashi D, Nomoto S, Kodera Y, et al. Suppressor of Cytokine Signaling 4 Detected as a Novel Gastric Cancer Suppressor Gene using Double Combination Array Analysis. World J Surg 2012;36:362-72. [Crossref] [PubMed]
- Guo W, Li W, Yuan L, et al. MicroRNA-106a-3p Induces Apatinib Resistance and Activates Janus-Activated Kinase 2 (JAK2)/Signal Transducer and Activator of Transcription 3 (STAT3) by Targeting the SOCS System in Gastric Cancer. Med Sci Monit 2019;25:10122-8. [Crossref] [PubMed]
- Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest 2007;117:2713-22. [Crossref] [PubMed]
- Chen Y, Wang D, Peng H, et al. Epigenetically upregulated oncoprotein PLCE1 drives esophageal carcinoma angiogenesis and proliferation via activating the PI-PLCepsilon-NF-kappaB signaling pathway and VEGF-C/ Bcl-2 expression. Mol Cancer 2019;18:1. [Crossref] [PubMed]
- Huang H, Wei L, Qin T, et al. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-kappaB signals. Cancer Biol Ther 2019;20:73-80. [Crossref] [PubMed]
- Li RC, Ke S, Meng FK, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis 2018;9:838. [Crossref] [PubMed]
- Yong YH, Wang P, Jia RM, et al. SOCS3 control the activity of NF-kappaB induced by HSP70 via degradation of MyD88-adapter-like protein (Mal) in IPEC-J2 cells. Int J Hyperthermia 2019;36:151-9. [Crossref] [PubMed]
- Li F, Zhang Z, Wang P, et al. ALC1 knockdown enhances cisplatin cytotoxicity of esophageal squamous cell carcinoma cells by inhibition of glycolysis through PI3K/Akt pathway. Life Sci 2019;232:116679. [Crossref] [PubMed]
- Cao J, Lv W, Wang L, et al. Ricolinostat (ACY-1215) suppresses proliferation and promotes apoptosis in esophageal squamous cell carcinoma via miR-30d/PI3K/AKT/mTOR and ERK pathways. Cell Death Dis 2018;9:817. [Crossref] [PubMed]
- Ma S, Liu T, Xu L, et al. Histone deacetylases inhibitor MS-275 suppresses human esophageal squamous cell carcinoma cell growth and progression via the PI3K/Akt/mTOR pathway. J Cell Physiol 2019;234:22400-10. [Crossref] [PubMed]
- Meng X, Dong X, Wang W, et al. Natural Borneol Enhances Paclitaxel-Induced Apoptosis of ESCC Cells by Inactivation of the PI3K/AKT. J Food Sci 2018;83:1436-43. [Crossref] [PubMed]
- Ha YNE, Dai Y, Wufuer D, et al. Second-generation Src/Abl inhibitor bosutinib effectively induces apoptosis in human esophageal squamous cell carcinoma (ESCC) cells via inhibiting Src/Abl signaling. Neoplasma 2020;67:54-60. [Crossref] [PubMed]
- Song M, Yoon G, Choi JS, et al. Janus kinase 2 inhibition by Licochalcone B suppresses esophageal squamous cell carcinoma growth. Phytother Res 2020;34:2032-43. [Crossref] [PubMed]
- Huang J, Li J, Li Y, et al. Interferon-inducible lncRNA IRF1-AS represses esophageal squamous cell carcinoma by promoting interferon response. Cancer Lett 2019;459:86-99. [Crossref] [PubMed]
- Zhang N, Zhang M, Wang Z, et al. Activated STAT3 Could Reduce Survival in Patients with Esophageal Squamous Cell Carcinoma by Up-regulating VEGF and Cyclin D1 Expression. J Cancer 2020;11:1859-68. [Crossref] [PubMed]
- Meng P, Wei J, Geng Y, et al. Targeted sequencing of circulating cell-free DNA in stage II-III resectable oesophageal squamous cell carcinoma patients. BMC Cancer 2019;19:818. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


