
REGγ promotes mantle cell lymphoma cell apoptosis by downregulating NF-κB signaling
Highlight box
Key findings
• The proteasome activator REGγ inhibits mantle cell lymphoma (MCL) cell proliferation by promoting cell apoptosis. REGγ inhibits MCL cell growth in mice by promoting apoptosis. REGγ-overexpression induces IκB accumulation and inhibits Nuclear factor-kappa B (NF-κB) signaling. REGγ controls NF-κB signal via STAT3-PSMB5 axis.
What is known and what is new?
• With the rapid development of molecular biology, targeted drugs of MCL have improved outcomes of conventional chemotherapy while avoiding long-term toxicities. The proteasome activator REGγ is implicated in the progression of cancer. As a promoter of cellular growth and a key regulator of several tumor suppressors, REGγ-proteasome as a potential target of new cancer-drug development.
• We found that REGγ regulated p-STAT3 expression by accelerating its half-life and downregulated the NF-κB signaling pathway to promote MCL cell apoptosis by negatively regulating STAT3-mediated PSMB5 expression and subsequently upregulating IκB expression.
What is the implication, and what should change now?
• Our discovery of the mechanism of REG in MCL provides a potential therapeutic target for its treatment.
Introduction
Mantle cell lymphoma (MCL) is an aggressive B-cell non-Hodgkin lymphoma (NHL), which accounts for 4–9% of NHLs worldwide (1). MCL is characterized by hallmark features such as t (11; 14) (q13; q32) chromosomal translocation and over-expression of cyclinD1, which lead to a poor prognosis (2). With the rapid development of molecular biology, targeted drugs such as bortezomib, ibrutinib, and lenalidomide have improved outcomes of conventional chemotherapy while avoiding long-term toxicities (3). However, MCL remains refractory to treatment owing to drug resistance. Therefore, it is necessary to identify new therapeutic targets.
Despite impressive achievements in targeted drugs, MCL has remained refractory to treatment due to tumoral resistance. The proteasome activator REGγ (also known as PSME3, PA28γ, and Ki antigen) is implicated in the progression of cancer. As a promoter of cellular growth and a key regulator of several tumor suppressors, a recent study has linked REGγ overexpression with tumor formation and suggested the REGγ-proteasome as a potential target of new cancer-drug development (4). REGγ is a member of the 11S proteasome. REGγ is present constitutively within the cell nucleus and is responsible for the degradation of most intracellular proteins in an ATP- and ubiquitin-independent manner by binding to and activating the 20S proteasome (5,6). REGγ is overexpressed in breast (7), thyroid (8) and colorectal cancers (9,10) and is correlated with poor prognosis in patients with MCL (6,11). However, the molecular mechanisms and specific roles of REGγ in MCL are unclear.
Previous studies demonstrated that REGγ promotes cell proliferation and migration and inhibits cell apoptosis by downregulating the nuclear factor-kappa B (NF-κB) signaling pathway in bowel inflammation (12), colon cancer (10), and multiple myeloma cells (13). Moreover, REGγ promotes inflammation in testicular Leydig cells via IκBε signaling (14). However, studies on the functional role and molecular mechanisms of REGγ in MCL remain scarce.
In this study, the JEKO-1 cell line was used to construct stable REGγ-knockdown and REGγ-overexpressing cell lines, and the effects and mechanisms of REGγ in MCLs were investigated. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2045/rc).
Methods
Animals and cell culture
All animals were purchased from Yongnuo Biotechnology Co. Ltd. (Guangzhou, China). Human JEKO-1 and REC-1 cells (American Type Culture Collection (ATCC), Rockefeller, MD, USA) were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) in an incubator at 37 ℃ and 5% CO2. Liposome 2000 reagent (Invitrogen, Carlsbad, CA, USA) was used to transfect cells in log phase growth. Experiments were performed under a project license (No. 2021-KZ-170-01) granted by Guangdong Second Provincial General Hospital Ethics Committee, in compliance with China’s national or institutional guidelines for the care and use of animals.
Cell transfection and treatment
In the presence of doxycycline (Dox, Pfizer Inc., New York, NY, USA), JEKO-1 cells stably transfected with the Tet-On system (CloneTech, Palo Alto, CA, USA) expressed high levels of REGγ. pLXSN-REGγ and pEGFP-N1 plasmids were digested with XhoI and XmaI restriction endonucleases, ligated with T4 ligase, and transformed into E. coli DH5α cells. The pEGFP-N1-REGγ plasmid was digested with XhoI and SalI, and pRevTRE was digested with SalI, ligated with T4 ligase, and transformed into E. coli DH5α cells. Plasmid DNA was then circularized with T4 ligase and transformed into DH5α cells to obtain pRevTRE-REGγ. pRevTet-on, pRevTRE, and pRevTRE-REGγ were transfected individually into the viral packaging cell line PT67 using polyethylenimine reagent. Stable cell lines PT67 pRevTRE and pRevTRE-REGγ were obtained through selection with G418 and hygromycin. The supernatant containing the virus was concentrated, purified, and then used to infect JEKO-1 cells. JEKO-1 cells with high REGγ expression were obtained by selection with G418 for 24 h.
Construction of stably transfected REGγ-knockdown cells using short hairpin RNA (shRNA) against REGγ. The special complementation fragments of REGγ (shN, 5'-UUCUCCGAACGUGUCACGUTT-3'; shREGγ, 5'-CAGAAGACUUGGUGGCAAA-3') were constructed and inserted into the lentiviral vector pHBLV-u6-puro (Huada, Guangzhou, China). Then, the lentiviral viruses were packaged by transfecting 293T cells for 24 h. Then, the obtained stably transfected cells were selected by treatment with 1 µg/mL of puromycin. The transfection efficiency of shRNA-REGγ on JEKO-1 and REC-1 cells was determined using western blotting.
Cell proliferation assay
The Cell Counting Kit 8 (CCK-8, Beyotime, Beijing, China) assay was used to detect JEKO-1 cell proliferation. First, JEKO-1 cells were seeded in 96-well plates (100 µL, 2,000 cells per well) at different time points after transfection, and then 10 µL CCK-8 solution was added to each well. The optical density of each well at 450 nm was measured using a spectrophotometer (Bio-Rad, Hercules, CA, USA) after 4 h of incubation.
Cell apoptosis assay
After transfection, JEKO-1 cell apoptosis was measured using a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, JEKO-1 cells in each group were suspended in 1× binding buffer 48 h after transfection. The cells were then stained with 5 µL Annexin V (Kegen, Nanjing, China, KGA1022) and 7 µL 7-AAD reagent (Invitrogen, MA, USA, 2115592). After incubation in the dark for 15 min, apoptosis levels were measured using a flow cytometer.
Western blot assays
The total proteins of JEKO-1 and REC-1 cells were leached using a lysis buffer. After centrifugation (12,000 rpm) at 4 ℃ for 15 min, 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the proteins. Proteins were then transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Membranes were blocked using 5% skim milk for 1 h at room temperature, and the specific diluted primary antibodies (REGγ, Caspase3, Cleaved Caspase3, PARP, Cyclin D1, BCL-2, Tubulin, p-NF-κB, NF-κB, IκBε, IκBβ, IκBα, p-STAT3, STAT3, and PSMB5) were incubated with the membranes overnight at 4 ℃. The membranes were washed three times with tris-buffered saline containing 0.1% Tween 20 solution, and then, horse radish peroxidase-conjugated secondary antibody (1:5,000, Abcam, Waltham, MA, USA) was incubated with the membranes for 1 h at 25 ℃. Finally, the protein bands were visualized using a ultra-violet (UV) gel imager (FluorChem M, Globalebio, Beijing, China). Cells were treated with or without STAT3 inhibitor Stattic (HY-13818, MCE, Shanghai, China) and then collected, and the protein expression of the NF-κB signaling pathway was detected by western blot assays. Separately, JEKO-1 cells were treated with cycloheximide (1 mg/mL) for 3–6 h, and the half-life of p-STAT3 was detected by western blot assays.
TUNEL assay
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays were used to evaluate cell apoptosis. First, the cells grown on coverslips were fixed with 4% paraformaldehyde, followed by washing with phosphate-buffered saline. Second, cells were treated with 0.3% H2O2 to inhibit the activity of endogenous peroxidase and then with the TUNEL reaction solution (Sigma-Aldrich, Merck KGaA, Germany) for 1 h at 37 ℃. Finally, they were imaged using a laser confocal microscope (Nikon A1 confocal microscope, Nikon Co., Japan).
Statistical analyses
All experiments were repeated three times. The data are presented as mean ± standard deviation (SD) and were analyzed using SPSS version 17.0 (SPSS Inc., Chicago, USA). The differences between two groups were compared using a t-test and those between more than three groups were compared using one-way analysis of variance (ANOVA). Differences were considered statistically significant at P values less than 0.05.
Results
REGγ inhibits MCL cell proliferation by promoting cell apoptosis
To assess the effect of REGγ on JEKO-1 cells, we constructed stable REGγ-knockdown (shR) and REGγ-overexpressing (+Dox) cell lines. Reducing levels of REGγ significantly promoted cell proliferation in a time-dependent manner (P<0.01), and higher expression of REGγ suppressed the proliferation of JEKO-1 cells (Figure 1A,1B) (P<0.01). Furthermore, the flow cytometry assay showed that the apoptotic rate was significantly lower in the shR JEKO-1 cells (P<0.001, Figure 1C) and significantly higher in the +Dox JEKO-1 cells than that in their corresponding controls (P<0.001, Figure 1D). These results indicate that silencing REGγ may efficiently promote cell proliferation by inhibiting apoptosis of JEKO-1 cells, and the higher expression of REGγ suppressed the proliferation of JEKO-1 cells by inducing apoptosis in vitro. However, flow cytometry assays revealed that REGγ had no effect on the cell cycle of MCL cells (Figure S1).
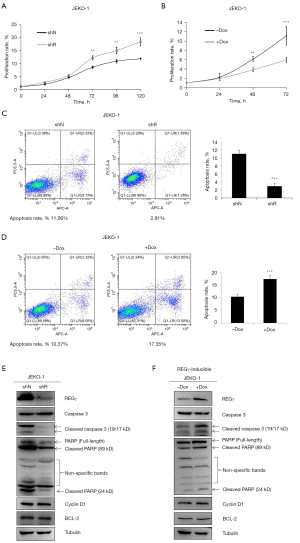
To further investigate the molecular mechanisms underlying the effects of REGγ on the cell cycle and apoptosis, we investigated the expression of proteins related to the cell cycle and apoptosis in shR and +Dox JEKO-1 cells. The silencing of REGγ in JEKO-1 reduced Caspase3-PARP-mediated apoptosis (Figure 1E). In contrast, higher expression of REGγ promoted Caspase3-PARP-mediated apoptosis (Figure 1F) and REGγ had no effect on cyclin D1 and BCL2 expression (Figure S2).
REGγ inhibits MCL cell growth in mice by promoting apoptosis
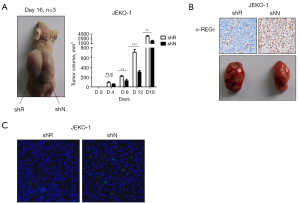
Based on our finding that REGγ inhibits MCL cell proliferation by promoting apoptosis in vitro, we next investigated the influence of REGγ on apoptosis in vivo. ShR and control (shN) JEKO-1 cells were inoculated subcutaneously into the flanks of nude mice, and tumor volume was measured every four days. We observed variability in tumor volumes generated from shR and shN JEKO-1 cells beginning on day 8 after the appearance of the tumors, and tumors generated from shR JEKO-1 cells grew more rapidly than tumors generated from shN JEKO-1 cells (P<0.01, Figure 2A,2B). The TUNEL assay was used to detect apoptotic cells in xenograft tumor tissues, and the results showed that more apoptotic cells were observed in tumor tissues in the shN group than in the shR group (Figure 2C).
REGγ-overexpression induces IκB accumulation and inhibits NF-κB signaling
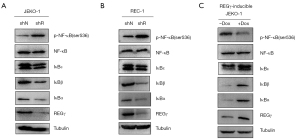
To explore the underlying mechanisms of REGγ-mediated MCL cell (JEKO-1 and REC-1) apoptosis, the protein expressions of key molecules involved in the NF-κB signaling pathway were investigated. Western blot assays revealed decreased expressions of the three types of IκB subunits and significantly increased levels of NF-κB in the shR group compared with those in the shN group (Figure 3A,3B), and increased expression of IκB and significantly decreased levels of NF-κB in the +Dox group compared with those in the control (−Dox) group (Figure 3C). NF-κB normally interacts with IκB, which leads to NF-κB being sequestered in the cytoplasm to inhibit its signaling (15). These results suggest that REGγ promotes MCL cell apoptosis by upregulating IκB expression to inhibit NF-κB activation. However, REGγ reduces IκBε and promotes NF-κB signaling in other cell types, including human lymphoma U937 cells, Burkitt lymphoma Namalwa cells, and mouse embryonic fibroblasts (Figure S3). These results indicate that the regulation of NF-κB and IκB expression by REGγ is cell-specific.
REGγ controls NF-κB signal via STAT3-PSMB5 axis
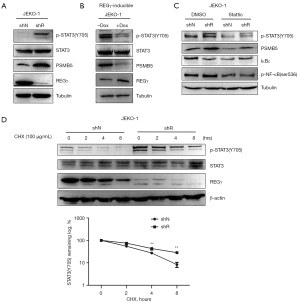
STAT3 is an important molecule in cancer therapy, as it interacts with the NF-κB signaling pathway and promotes tumor progression (16-18). PSMB5 is the molecular target of bortezomib, used clinically to ameliorate relapsed multiple myeloma and MCL. Activated STAT3 levels in cancers may presumably circumvent the effect of bortezomib regimen through up-regulation of PSMB5 protein. These observations suggest that STAT3 modulates PSMB5 expression in MCL (19). To further elucidate the specific molecular mechanism by which REGγ regulates the NF-κB pathway, we examined STAT3 and PSMB5 expression in MCL cells with overexpressed and silenced REGγ. The results revealed that REGγ knockdown activated STAT3 and upregulated PSMB5 expression (Figure 4A), whereas REGγ overexpression inhibited STAT3 and downregulated PSMB5 expression in JEKO-1 cells (Figure 4B). We explored the role of REGγ in the STAT3-PSMB5 signaling axis regulating NF-κB signaling pathways by treating shN and shR JEKO-1 cells with the STAT3 inhibitor Stattic. The results demonstrated that Stattic downregulated PSMB5 expression, upregulated IκBε expression, and downregulated NF-κB expression in JEKO-1 cells, in both shN and shR cells (Figure 4C). We found that REGγ is involved in the regulation of p-STAT3 levels by accelerating its half-life (Figure 4D). Taken together, these results show that REGγ regulates NF-κB signaling by downregulating STAT3-mediated PSMB5 expression, subsequently influencing the expression of IκB.
Discussion
REGγ is important for tumor occurrence and development, but understanding of the specific role of REGγ in MCL is lacking. In this study, we found that REGγ inhibits MCL cell proliferation by promoting cell apoptosis, and the downregulated NF-κB signaling is closely related to the regulation of apoptosis. But REGγ reduces IκBε and promotes NF-κB signaling in other cell types (human lymphoma U937 cells, Burkitt lymphoma Namalwa cells, and mouse embryonic fibroblasts). REGγ downregulated PSMB5 expression by affecting the level of p-STAT3, accelerating its half-life, upregulating IκB expression, and subsequently inhibiting NF-κB signaling to promote MCL cell apoptosis.
REGγ is a member of the 11S proteasome and is upregulated in several types of cancers, such as thyroid, breast, lung, colorectal, and liver cancers (11,20). In previous reports, the overexpression of REGγ at the cellular level not only promoted cell proliferation and migration of lung cancer cells but also increased the number of cells in the S and G2/M phases (21). Silencing of REGγ suppressed cell proliferation, migration, and invasiveness of endometrial cancer cells (22). Surprisingly, our findings revealed the opposite result in MCL cells. We found that REGγ inhibits MCL cell proliferation by promoting apoptosis, suggesting that upregulation of REGγ may be a potential therapeutic strategy for MCL. In addition, REGγ had no effect on the MCL cell cycle.
The NF-κB signaling pathway plays a crucial role in the proliferation and apoptosis of MCL cells. Thus, targeting the NF-κB pathway might be a potential approach in treating MCL (23,24). In this study, REGγ knockdown significantly increased the expression of IκB and decreased the levels of NF-κB. The results clearly indicate that silencing REGγ resulted in decreased expression of the three types of IκB subunits and significantly increased levels of NF-κB. Upregulation of IκB expression causes inhibition of the NF-κB signaling pathway, leading to reduced proliferation and increased apoptosis of MCL cells (25). These studies agree with our findings, which further show that the inhibitory effect of REGγ on MCL cells is closely related to the downregulation of the NF-κB signaling. We found that REGγ decreased IκBε and promoted NF-κB signaling in other cell types, which indicates that the regulation of NF-κB and IκB expression by REGγ is cell-specific.
STAT3 plays a crucial role in the formation and metastasis of cancer, as well as the development of drug resistance (6,11,20). In tumor cells, STAT3 often interacts with NF-κB to advance tumor progression (16-18). Additionally, it has been suggested that STAT3 could potentially have a role in the biological function of MCL (26). In this study, REGγ knockdown activated STAT3 and upregulated PSMB5 expression, whereas REGγ overexpression inhibited STAT3 and downregulated PSMB5 expression in JEKO-1 cells. Treating the shN and shR JEKO-1 cells with the Stattic downregulated PSMB5 expression, upregulated IκBε expression, and downregulated NF-κB expression in JEKO-1 cells, independent of REGγ expression, suggesting that REGγ regulates NF-κB signaling pathways to promote the apoptosis of JEKO-1 cells by downregulating STAT3-mediated PSMB5 expression and subsequently influencing the expression of IκB. Meanwhile, p-STAT3 degradation in shR JEKO-1 cells was slower than that in the control group, indicating that REGγ is involved in the regulation of p-STAT3 expression by accelerating its half-life. Previous research has established that STAT3 knockdown significantly reduces expression of the transcription factor PSMB5, and IκB expression is negatively regulated by PSMB5 (18). Collectively, our results further suggest that REGγ inhibits NF-κB signaling pathways to promote apoptosis of MCL cell by inhibiting STAT3 activity through shortening half-life of p-STAT3, which downregulates PSMB5 expression and subsequently influences the expression of IκB. However, several studies have indicated that REGγ promotes tumorigenesis and the development of cancers by promoting degradation of target proteins, which regulate multiple signaling pathways (27-29). This is in contrast to our present findings indicating that REGγ inhibits the growth and proliferation of MCL cells.
Conclusions
REGγ may inhibit cell proliferation while promoting apoptosis of MCL cells by downregulating the NF-κB signaling pathway. In parallel, this process is potentially highly relevant to the downregulation of STAT3 and PSMB5 expressions and upregulation of IκBε expression in MCL cells. However, the present study was mainly limited to specific cells and one animal model. The mechanism of action of REGγ in MCL still requires further investigation.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2045/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2045/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2045/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2045/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. 2021-KZ-170-01) granted by Guangdong Second Provincial General Hospital Ethics Committee, in compliance with China’s national or institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pararajalingam P, Coyle KM, Arthur SE, et al. Coding and noncoding drivers of mantle cell lymphoma identified through exome and genome sequencing. Blood 2020;136:572-84. [Crossref] [PubMed]
- Balaji S, Ahmed M, Lorence E, et al. NF-κB signaling and its relevance to the treatment of mantle cell lymphoma. J Hematol Oncol 2018;11:83. [Crossref] [PubMed]
- Cliff ERS, Dickinson M. Treatment of Mantle-Cell Lymphoma. N Engl J Med 2022;387:1146-7. [Crossref] [PubMed]
- Funderburk KE, Kang J, Li HJ. Regulation of Life & Death by REGγ. Cells 2022;11:2281. [Crossref] [PubMed]
- Gao X, Chen H, Liu J, et al. The REGγ-Proteasome Regulates Spermatogenesis Partially by P53-PLZF Signaling. Stem Cell Reports 2019;13:559-71. [Crossref] [PubMed]
- Mao I, Liu J, Li X, et al. REGgamma, a proteasome activator and beyond? Cell Mol Life Sci 2008;65:3971-80. [Crossref] [PubMed]
- Yi Z, Yang D, Liao X, et al. PSME3 induces epithelial-mesenchymal transition with inducing the expression of CSC markers and immunosuppression in breast cancer. Exp Cell Res 2017;358:87-93. [Crossref] [PubMed]
- Bhatti MZ, Pan L, Wang T, et al. REGγ potentiates TGF-β/Smad signal dependent epithelial-mesenchymal transition in thyroid cancer cells. Cell Signal 2019;64:109412. [Crossref] [PubMed]
- Song W, Guo C, Chen J, et al. Silencing PSME3 induces colorectal cancer radiosensitivity by downregulating the expression of cyclin B1 and CKD1. Exp Biol Med (Maywood) 2019;244:1409-18. [Crossref] [PubMed]
- Wang Q, Gao X, Yu T, et al. REGγ Controls Hippo Signaling and Reciprocal NF-κB-YAP Regulation to Promote Colon Cancer. Clin Cancer Res 2018;24:2015-25. [Crossref] [PubMed]
- He J, Cui L, Zeng Y, et al. REGγ is associated with multiple oncogenic pathways in human cancers. BMC Cancer 2012;12:75. [Crossref] [PubMed]
- Xu J, Zhou L, Ji L, et al. The REGγ-proteasome forms a regulatory circuit with IκBɛ and NFκB in experimental colitis. Nat Commun 2016;7:10761. [Crossref] [PubMed]
- Liu S, Zheng LL, Zhu YM, et al. Knockdown of REGγ inhibits the proliferation and migration and promotes the apoptosis of multiple myeloma cells by downregulating NF-κB signal pathway. Hematology 2018;23:277-83. [Crossref] [PubMed]
- Xie T, Chen H, Shen S, et al. Proteasome activator REGγ promotes inflammation in Leydig cells via IkBε signaling. Int J Mol Med 2019;43:1961-8. [Crossref] [PubMed]
- Mussbacher M, Salzmann M, Brostjan C, et al. Cell Type-Specific Roles of NF-κB Linking Inflammation and Thrombosis. Front Immunol 2019;10:85. [Crossref] [PubMed]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009;9:798-809. [Crossref] [PubMed]
- Fan Y, Mao R, Yang J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013;4:176-85. [Crossref] [PubMed]
- Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol 2018;18:309-24. [Crossref] [PubMed]
- Vangala JR, Dudem S, Jain N, et al. Regulation of PSMB5 protein and β subunits of mammalian proteasome by constitutively activated signal transducer and activator of transcription 3 (STAT3): potential role in bortezomib-mediated anticancer therapy. J Biol Chem 2014;289:12612-22. [Crossref] [PubMed]
- Chai F, Liang Y, Bi J, et al. High expression of REGγ is associated with metastasis and poor prognosis of patients with breast cancer. Int J Clin Exp Pathol 2014;7:7834-43. [PubMed]
- Qin Q, Guo FC, Luo ST, et al. REGgamma promotes malignant behaviors of lung cancer cells. Sichuan Da Xue Xue Bao Yi Xue Ban 2014;45:304-8. [PubMed]
- Wang H, Bao W, Jiang F, et al. Mutant p53 (p53-R248Q) functions as an oncogene in promoting endometrial cancer by up-regulating REGγ. Cancer Lett 2015;360:269-79. [Crossref] [PubMed]
- Gao X, Wang Q, Wang Y, et al. The REGγ inhibitor NIP30 increases sensitivity to chemotherapy in p53-deficient tumor cells. Nat Commun 2020;11:3904. [Crossref] [PubMed]
- Roué G, Sola B. Management of Drug Resistance in Mantle Cell Lymphoma. Cancers (Basel) 2020;12:1565. [Crossref] [PubMed]
- Wang T, Fahrmann JF, Lee H, et al. JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab 2018;27:136-150.e5. [Crossref] [PubMed]
- Vogt N, Dai B, Erdmann T, et al. The molecular pathogenesis of mantle cell lymphoma. Leuk Lymphoma 2017;58:1530-7. [Crossref] [PubMed]
- Li X, Lonard DM, Jung SY, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell 2006;124:381-92. [Crossref] [PubMed]
- Gao G, Wong J, Zhang J, et al. Proteasome activator REGgamma enhances coxsackieviral infection by facilitating p53 degradation. J Virol 2010;84:11056-66. [Crossref] [PubMed]
- Ali A, Wang Z, Fu J, et al. Differential regulation of the REGγ-proteasome pathway by p53/TGF-β signalling and mutant p53 in cancer cells. Nat Commun 2013;4:2667. [Crossref] [PubMed]


