
miR-17 enhances proliferation and migration and inhibits apoptosis in glioma cells by regulating SASH1 expression
Introduction
Glioma is the most common intracranial malignant tumor, accounting for approximately 35–61% of intracranial tumors (1,2). Gliomas also display extremely high invasiveness and infiltration rates and are one of the most difficult types of tumors to cure (3). Improvements in neuroimaging techniques and microsurgical procedures have improved the surgical treatment of glioma, and recent molecular biology studies are pointing the way towards potential gene therapy tools; however, to date, there have been no significant breakthroughs in the treatment of this disease (4,5). The most comprehensive current treatment regimen for glioma consists of surgical treatment, radiotherapy, and medication (6-8). Therefore, a pressing problem in the field of glioma neurosurgery is how to optimize existing measures and create new methods to enhance treatment efficacy, extend patient survival time, and increase quality of life (9,10).
The discovery of microRNAs (miRNAs) has opened up new paths for studying the molecular mechanisms underlying the occurrence and development of gliomas as well as for offering new possibilities for therapeutic tools (11,12). miRNAs are a group of small non-coding RNA molecules 18–25 nucleotides in length, and they play important roles in many evolutionarily conserved processes (13,14). miRNAs primarily function at the post-transcription level as negative regulators of gene expression. However, their precise mode of action depends on the degree of complementarity between the miRNA and its target mRNA. In particular, full complementarity with a sequence in the 3’-untranslated region (3’-UTR) of its target gene can lead to mRNA degradation or cutting, similar to the mechanism of small interfering RNA (siRNA) silencing (15,16). Furthermore, individual miRNAs can regulate the mRNAs of many different target genes, and individual target mRNAs can be regulated by multiple miRNAs (17,18).
miRNAs exert their effects primarily through the inhibition or promotion of target gene expression (19,20). miRNAs control the temporal and spatial expression of many genes in a variety of developmental contexts in animals, including histogenesis, organogenesis (e.g., nerve, muscle, and blood development), cell physiology (e.g., apoptosis and metabolism), and signaling pathways (14,20). In addition, the association between miRNAs and tumors has become an important part of cancer research (21,22). Many studies have demonstrated that miRNAs are closely linked with the occurrence, development, and therapeutic reactions of tumors, and they are expected to serve as ideal molecular markers and targets for tumor diagnosis, prognosis and treatment (23,24). Indeed, in the emerging field of “oncomirs” and other tumor-related miRNAs, studies describing associations between single miRNAs or miRNA gene clusters and tumor occurrence are continuously published. One of the most intensively studied miRNA gene clusters is the human microRNA-17-92 (miR-17-92) cluster. This cluster is composed of 6 miRNAs (miR-17-5p, miR-18a, miR-19a, miR-20, miR-19b-1, and miR-92-1) and is located on chromosome 13q31.3 in the C13orf25 open reading frame (25,26). This locus shows abnormal expression in many malignant tumors, including diffuse B-cell lymphoma, lung cancer, and Burkitt’s lymphoma (27,28). To date, increased expression of members of this gene cluster has been reported in a variety of solid tumors, including colon cancer, prostate cancer, thyroid cancer, lung cancer, and malignant blood tumors. Functional studies have shown that miR-17-92 can participate in tumor development by promoting cell cycle progression, cell proliferation, and angiogenesis as well as by inhibiting apoptosis (29-31). However, reports on the effects of miR-17 in glioma cells are relatively rare.
SAM- and SH3-domain containing 1 (SASH1) is a newly discovered tumor suppressor gene (32). The SASH1 gene is located on 6q24.3 and contains 22 exons and 21 introns. Zeller et al. first reported that SASH1 acts as a tumor suppressor in breast cancer (32), and in 2006, Rimkus et al. reported that SASH1 also acts as a tumor suppressor in colon cancer (33). Previous studies by our group revealed that SASH1 gene expression can regulate the inhibition of growth, proliferation, and invasion in U251 cells as well as the promotion of apoptosis in this cell type (34). In addition, using on-line TargetScan software analysis, we showed that SASH1 may be a target of miR-17. Therefore, we hypothesized that miR-17 functions as a tumor-promoting miRNA through inhibition of the SASH1 tumor suppressor gene in glioma cells.
In the present study, we analyzed the expression of miR-17 in human glioma tissues and cells and determined its effects on various cell biological characteristics in glioma cells. We demonstrate that miR-17 can inhibit the expression of SASH1, suggesting new strategies for the treatment of human gliomas.
Methods
Specimens
A total of 20 cases of fresh specimens resected from glioma patients treated in our hospital and 6 cases of normal adjacent tissues were collected and stored immediately in liquid nitrogen. None of the patients had a history of preoperative treatment, including chemotherapy and radiotherapy. None of the patients had other inflammatory diseases.
Major reagents
HEK293 cells, U251 and SHG-44 human glioma cell lines, and the C6 rat glioma cell line were purchased from ATCC (USA). Fetal bovine serum (FBS), DMEM, RPMI 1640, L-glutamine, HEPES, and LipofectamineTM 2000 were purchased from Invitrogen (San Diego, CA, USA). Cell culture plates, dishes, and Transwell migration chambers were purchased from Corning (Corning, NY, USA). Matrigel was purchased from BD Bioscience (San Jose, CA, USA).
The TaqMan miRNA Isolation Kit, TaqMan microRNA Assay kit, TaqMan microRNA Assay and TaqMan Universal PCR Master Mix were purchased from Applied Biosystems (USA).
MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide] was purchased from Sigma Aldrich (USA). The propidium iodide (PI) FITC-Annexin V apoptosis detection kit was purchased from BD (USA). The pGL3-Luciferase plasmid and luciferase detection kit were purchased from Promega (USA). The miR-17 inhibitor, inhibitor control, miR-17 mimic and mimic control were synthesized by GenePharma (Shanghai, China).
Hoechst33342, RIPA lysis buffer, and TBST (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween-20) were purchased from Beyotime Biotech (Shanghai, China). The Bradford protein concentration determination kit and the QuantityOne software program were purchased from Bio-Rad (Richmond, CA, USA). PVDF membrane was purchased from Millipore (Bedford, MA, USA). The ECL chemiluminescent reagent kit was purchased from Pierce (Rockford, Illinois, USA).
The rabbit anti-human SASH1 polyclonal antibody and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG polyclonal antibody were purchased from Novus Biologicals (Littleton, CO, USA). The rabbit anti β-actin polyclonal antibody was purchased from Abbiotec Corp. (San Diego, CA, USA).
Cell treatment
The Human U251 glioma cell line, rat C6 glioma cell line, and HEK293 cells were cultured in DMEM containing 10% FBS. The SHG-44 human glioma cell line was cultured in RPMI 1640 containing 10% FBS. Culturing was performed at 37 °C under 5% CO2 and saturated humidity conditions. The growth status of the cells was observed using an inverted microscope (Leica DM IL LED, German). When the cells reached 70–80% confluence, cells were passaged by dissociation with 0.25% trypsin. The culture medium was changed for every other day, and passaging was conducted for every 3–4 days. Cells in the logarithmic phase were collected for experiments.
Normally cultured U251 cells were evenly inoculated onto 96- or 6-well plates at 3×105 cells/mL with 100 or 1,000 µL in each well, respectively. Transfection of the miR-17 inhibitor and the negative control (inhibitor control) was performed according to the Lipofectamine 2000 transfection manual; a normal control group was also set up. The miR-17 inhibitor and the inhibitor control were diluted in serum-free MEM medium. Lipofectamine 2000 liposomes were diluted in MEM medium, mixed gently, and incubated at room temperature for 5 min. The diluted Lipofectamine 2000 and the diluted miR-17 inhibitor or inhibitor control were mixed gently and incubated at room temperature for 20 min to form complexes. The complexes were added to the culture plates containing U251 cells and cultured in an incubator at 37 °C under 5% CO2. After 5 h, the culture medium was replaced with MEM culture medium containing 10% FBS and the cells were cultured for another 48 h.
Detection of miR-17 expression in human glioma tissues and cell lines using real-time quantitative PCR (qRT-PCR)
miR-17 expression in human glioma tissues, adjacent normal tissues, and different glioma cell lines (U251, SHG-44, and C6) was detected using qRT-PCR. Different in vitro cultured glioma cells and glioma tissues were collected, and RNA was extracted using the TaqMan miRNA separation kit. The expression of mature miR-17 was detected using the TaqMan microRNA Assay and TaqMan Universal PCR Master Mix; U6 was used as the internal control. All reactions were performed in three replicate wells. The cycle threshold (Ct) value of each reaction was recorded. Experimental results were analyzed using the relative quantitative method for qRT-PCR.
Quantifying the effects of miR-17 inhibitor transfection on miR-17 expression in U251 cells using qRT-PCR
The effects of miR-17 inhibitor transfection on miR-17 expression in U251 cells were detected using qRT-PCR. Transfection of the miR-17 inhibitor and the inhibitor control was conducted according to the Lipofectamine 2000 transfection manual; a normal control group was also set up. Transfections were conducted for 48 h. RNA was extracted using the TaqMan miRNA separation kit. The expression of miR-17 in each group was determined using the TaqMan microRNA Assay and TaqMan Universal PCR Master Mix.
Quantifying the effects of miR-17 on cell viability using the MTT assay
After U251 cells were transfected with the miR-17 inhibitor or the inhibitor control for 48 h, each well was treated with 100 µL of MTT (0.5 mg/mL) solution and incubated in a 37 °C under 5% CO2 incubator for 4 h. Each well was then treated with 20% SDS (co-solvent: 50% dimethylformamide) at 37 °C for 24 h; a normal control group was set up at the same time. The optical density (OD) value was measured at 570 nm using a microplate reader (Bio-Tek, USA). Each experimental group had 10 replicate wells, and the experiment was repeated 3 times.
Quantifying the effects of miR-17 on cell proliferation using flow cytometry
Normally cultured U251 cells were evenly inoculated into 6-well plates at 3×105 cells/mL in 1,000 µL per well. Transfection of the miR-17 inhibitor and the inhibitor control was performed according to the Lipofectamine 2000 transfection manual; a normal control group was also set up. After transfection for 48 h, the cells were washed 1–2 times with PBS and then dissociated with trypsin. After washing twice with PBS, the cells were then centrifuged and collected. PI staining solution was added for labeling at 4 °C for 30 min. After filtering through a screen mesh, cells were analyzed using a flow cytometer (BD). Cells were counted using the FCM CellQuest software program, and data were analyzed using the Macquit software program.
Quantifying the effects of miR-17 on cell apoptosis using flow cytometry
U251 cells were transfected with the miR-17 inhibitor or the inhibitor control; a normal control group was also set up. After transfection for 48 h, cells were washed 1–2 times with PBS, and Annexin V-FITC and PI staining solution were added for labeling in the dark for 15 min. After filtering through a screen mesh, cells were analyzed using a flow cytometer (BD). Cells were counted using the FCM CellQuest software program, and data were analyzed using the Mac quit software program.
Quantifying the effects of miR-17 on cell migration using the Transwell migration assay
Normally cultured U251 cells were evenly inoculated into 6-well plates at 3×105 cells/mL in 1,000 µL per well. Transfection of the miR-17 inhibitor and the inhibitor control was performed according to the Lipofectamine 2000 transfection manual; a normal control group was also set up. After 24 h of transfection, U251 cells were inoculated into a Transwell migration chamber and cultured for 24 h under normal conditions. Cells were washed 1–2 times with PBS, fixed in 4% paraformaldehyde, and stained with Hoechst33342. The unmigrated cells were removed with cotton swabs. The number of cells that passed through the polycarbonate membrane of the Transwell chamber was counted using a Leica microscope. Cells that passed through the Transwell polycarbonate membrane were classified as “invading” cells. A total of 8 randomly selected fields were quantified per sample.
Identification of SASH1 as a miR-17 target gene using a luciferase reporter gene system
To construct the luciferase plasmid, we chemically synthesized a fragment containing the SASHI 3’UTR sequence that is complementary to miR-17 as well as a mutant version of the SASH1 3’UTR. The SASH1 3’UTR fragment containing the predicted miR-17 binding site and the mutant SASH1 3’UTR fragment were cloned into the XbaI site of the PGL3-Luciferase vector.
Normally cultured HEK293 cells were evenly inoculated into 6-well plates at 3×105 cells/mL in 1,000 µL per well. We then cotransfected the SASH1 3’UTR luciferase plasmid and either the miR17 mimic or the mimic control; a normal control group was also set up. After transfection for 48 h, the cells were collected and tested using a luciferase reagent kit, and the results were analyzed using a microplate reader.
Quantifying the effects of miR-17 inhibitor transfection on SASH1 expression in U251 cells
Using the same transfection method described above, cellular protein was collected from each group after transfection for 48 h. Cellular SASH1 protein expression was detected using Western blots. In 6-well culture plates, each well was treated with 1 mL of RIPA lysis buffer [150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 7.9), 10 mM NaF, 10 mM PMSF and 1× protease inhibitors (Complete Cocktail Tablets, Roche)]. The cell lysis solution was transferred into a 1.5-mL centrifuge tube and centrifuged at 16,000 g for 30 min. The supernatant was collected, and protein concentration was determined using the BCA method. A 5% stacking gel and a 10% resolving gel were prepared, and each well was loaded with 50 µg of total protein. The proteins were separated by electrophoresis and then wet transferred onto a PVDF membrane (Bio-Rad, USA). The membrane was blocked in TBST (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween-20) containing 5% non-fat milk at room temperature for 1 h. The membrane was then incubated with rabbit anti-human SASH1 polyclonal antibodies (1:500 dilution) and rabbit anti-human β-actin polyclonal antibodies (1:1,000 dilution) at 4 °C overnight. After washing three times for 5 minutes with TBST, the membrane was incubated with HRP-labeled goat anti-rabbit IgG secondary antibodies at 37 °C for 1 h. After washing three times for five minutes with TBST, the membranes were developed with the ECL chemiluminescent reagent. The relative levels of SASH1 expression was determined using the gray density ratio of SASH1:β-actin. Relative expression changes were analyzed using the PDQuest software program (Bio-Rad, Richmond, CA, USA).
This study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University, and it was conformed to the provisions of the Declaration of Helsinki (as revised in Edinburgh 2000).
Statistical analysis
Statistical analyses were performed using the SPSS17.0 statistical software package. Comparisons between two groups were conducted using t-tests, and comparisons of data for more than two groups were conducted using analysis of variance. All experiments were repeated 3 times. P<0.05 indicates a statistically significant difference.
Results
Detection of miR-17 expression in human glioma tissues and cell lines using qRT-PCR
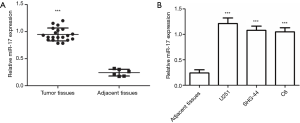
qRT-PCR results revealed that miR-17 expression in glioma tissues was significantly higher than that in normal adjacent tissues (P<0.001) (Figure 1A). In addition, miR-17 expression in glioma cell lines (U251, SHG-44, and C6) was significantly higher than in normal adjacent tissues (P<0.001) (Figure 1B).
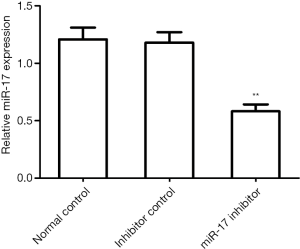
The effects of miR-17 inhibitor transfection on miR-17 expression in U251 cells
To inhibit miR-17 expression, the miR-17 inhibitor was transfected into U251 cells. The qRT-PCR results showed that miR-17 expression in the miR-17 inhibitor transfection group was significantly lower than in the normal control group (P<0.01) and in the inhibitor control group (P<0.01) (Figure 2).
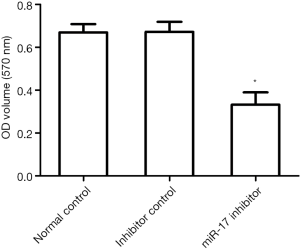
The effects of miR-17 on cell viability using the MTT assay
The MTT assay results showed that cell viability in the miR-17 inhibitor transfection group was significantly lower than in the normal control and inhibitor control groups (P<0.05) (Figure 3). These results indicate that inhibition of miR-17 expression reduces cell viability.
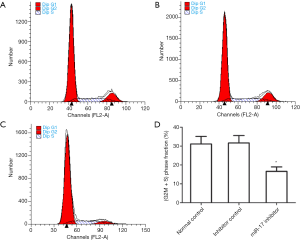
The effects of miR-17 on cell proliferation
After U251 cells were transfected with the miR-17 inhibitor or the inhibitor control for 48 h, changes in cell proliferation in each group were analyzed by flow cytometry. The fraction of cells in the G2M+S phase was set as equal to (G2M+S)/(G0G1+S+G2M). The results from the flow cytometry analysis indicated that the fraction of cells in the G2M+S phase in the miR-17 inhibitor transfection group was significantly lower than in the normal control group (P<0.05) and in the inhibitor control group (P<0.05) (Figure 4). These results indicate that inhibition of miR-17 expression can inhibit U251 cell proliferation.
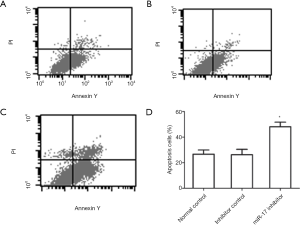
The effects of miR-17 on cell apoptosis
The flow cytometry results indicated that the percentage of apoptotic cells in the miR-17 inhibitor transfection group was significantly higher than in the normal control group (P<0.05) and in the inhibitor control group (P<0.05) (Figure 5).
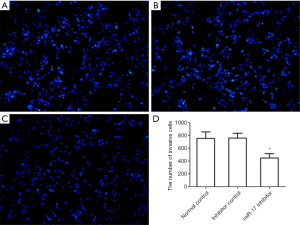
The effects of miR-17 on cell migration
After U251 cells were transfected with the miR-17 inhibitor or the inhibitor control for 48 h, Transwell migration chamber assays were performed to detect changes in the migratory ability of cells in each group. The results showed that after miR-17 inhibitor transfection, the number of cells that passed through the Transwell membrane was significantly decreased (P<0.05). These results indicate that transfection with the miR-17 inhibitor inhibits migration in U251 cells (Figure 6).
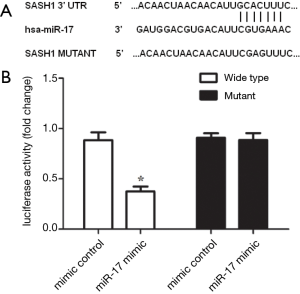
SASH1 is a miR-17 target gene
To validate that the predicted miR-17 target sequence in the SASH1 3’-UTR is functional, luciferase plasmids containing either the SASH1 3’UTR or a mutant version of the SASH1 3’UTR were constructed and co-transfected into HEK293 cells with either a miR-17 mimic or a mimic control. The results showed that luciferase activity in the miR-17 inhibitor transfection group was significantly decreased compared with the inhibitor control (P<0.05) (Figure 7). These results suggest that miR-17 directly binds to the predicted target site in the SASH1 3’-UTR.
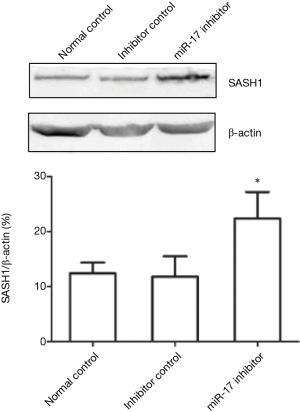
The effect of miR-17 inhibitor transfection on SASH1 expression in U251 cells
The western blot results showed that SASH1 protein expression in the miR-17 inhibitor transfection group was significantly higher than in the normal control and inhibitor control groups (P<0.05) (Figure 8). These results indicate that inhibition of miR-17 expression can promote SASH1 protein expression.
Discussion
Gliomas are tumors derived from neural epithelia, and they are characterized by high incidence, high recurrence, high mortality, and low cure rates (35-37). Malignant glioma refractory is mediated to a large extent by its strong invasive features. Currently, there are no highly effective radical treatment methods for this type of tumor, and the results of conventional treatment methods, such as surgical resection, chemotherapy, and radiotherapy, are far from ideal (38,39). Therefore, increasing glioma treatment efficacy is crucially important in the field of neurosurgery.
The miR-17-92 cluster is a typical polycistronic miRNA gene that is highly conserved in many species (40). The abnormally high expression of members of this gene cluster has been reported in a variety of solid tumors, including colon cancer, prostate cancer, thyroid cancer, lung cancer, and malignant blood tumors. Functional studies showed that miR-17-92 can participate in tumor development by promoting cell cycle progression, cell proliferation, and angiogenesis as well as by inhibiting apoptosis (29-31). The transfection of endothelial cells with the miR-17-92 cluster at the time of VEGF stimulation can partially inhibit the cell proliferation and morphogenetic changes initiated by the loss of Dicer. In addition, it has also been reported that miR-17-92 family members are associated with the proliferation and differentiation of lung progenitor cells (41). In this study, we used MTT, flow cytometry, and Transwell migration chamber experiments to validate the effects of miR-17 on the U251 glioma cell line. Our results revealed that cell viability, proliferation index, and invasiveness were all decreased in the miR-17 inhibitor transfection group, whereas apoptosis was significantly increased. These results indicate that miR-17 promotes viability, proliferation, and invasiveness and inhibits apoptosis in these tumors.
The SASH1 gene encodes a protein with both SH3 and SAM domains, both of which can mediate protein-protein interactions. The functions of the SAM domain are relatively complex, and the SAM domain can mediate both homologous and heterologous oligomerization through SAM domains in other proteins. Furthermore, SAM domains can also mediate interactions between the Smaug protein and mRNAs to mediate transcriptional regulation (42). Therefore, the fact that SASH1 contains these two structural domains suggests that this protein plays an important role in intracellular signal transduction (43-47). Indeed, existing studies on the SASH1 gene suggested that it can act as a tumor suppressor. However, studies on the mechanisms of SASH1 function are scarce. Lin et al. reported that the tumor suppressive activity of SASH1 is mediated by G2/M arrest in melanoma A-375 cells, which could be due to the phosphorylation of Cdc2 or the disruption of cyclin B-Cdc2 binding (48). In addition, Sheyu et al. reported increased DNA methylation levels in the promoter region of SASH1, particularly at the CpG_26.27 and CpG_54.55 sites, which could lead to the repression of SASH1 expression in breast cancer (49). Therefore, we speculated that in addition to the regulation of SASH1 by methylation, the inhibition of SASH1 in U251 glioma cells might also be regulated by miR-17.
Many miR-17 target genes have already been discovered, most of which have functions in cell proliferation, differentiation, apoptosis, and migration. The present study used a luciferase reporter gene system to show that SASH1 is a bona fide target gene of miR-17. Compared with the inhibitor control group, luciferase activity in the miR-17 inhibitor transfection group was significantly increased (P<0.05), demonstrating that miR-17 directly acts on the predicted target site in the SASH1 3’-UTR. The regulation of SASH1 protein expressed by miR-17 was also analyzed by Western blotting, further confirming that SASH1 is a functional target of this miRNA.
A study by Zhou et al. reported that SASH1 mutations induced the loss of E-Cadherin in human A375 cells, suggesting that SASH1 acts as a scaffolding protein to regulate IQGAP1-E-Cadherin signaling, and they also demonstrated novel cross talk between GPCR signaling and calmodulin signaling during the modulation of melanocyte invasion (50). Indeed, E-cadherin is closely associated with the migration of glioma cells (51). In addition, Carraro et al. reported that miR-17 can inhibit E-cadherin expression (52), and Li et al. reported that the inhibition of miR-17 can inhibit MMP-9 activity (53). In this study, we found that miR-17 promoted U251 cell invasion, and previous results from our group suggested that SASH1 can inhibit MMP-9 expression and inhibit U251 cell migration. Therefore, we speculate that miR-17 regulates the function of SASH1 in U251 to alter cell proliferation and migration by regulating MMP-9 and E-cadherin expression.
Sheyu et al. reported that SASH1 is regulated by methylation in breast cancer cells (49), and Dakhlallah et al. reported that the miR-17–92 miRNA cluster is critical for lung development and lung epithelial cell homeostasis by targeting the expression of fibrotic genes and DNA methyltransferase (DNMT)-1 (54). Therefore, whether miR-17 affects the methylation of the SASH1 promoter by regulating DNMT-1 expression requires further investigation.
In conclusion, miR-17 is up-regulated in human glioma tissues and cells lines. miR-17 negatively regulates SASH1 protein expression, promotes proliferation and invasiveness, and inhibits apoptosis in U251 glioma cells. Therapeutic strategies based on inhibiting miR-17 expression could benefit glioma patients.
Acknowledgments
Funding: This study was supported by grants from the Six Talent Peaks Project of Jiangsu Province (2014-WSW-027) and the Natural science foundation of Education Department of Jiangsu Province (11KJA180004).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.04.06). Liu Yang serves as an unpaid editorial board member of Translational Cancer Research from Jun 2012 - May 2019. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University, and it was conformed to the provisions of the Declaration of Helsinki (as revised in Edinburgh 2000) and the later amendment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ryken TC, Kalkanis SN, Buatti JM, et al. The role of cytoreductive surgery in the management of progressive glioblastoma: a systematic review and evidence-based clinical practice guideline. J Neurooncol 2014;118:479-88. [Crossref] [PubMed]
- Vigneswaran K, Neill S, Hadjipanayis CG. Beyond the World Health Organization grading of infiltrating gliomas: advances in the molecular genetics of glioma classification. Ann Transl Med 2015;3:95. [PubMed]
- Koshkin PA, Chistiakov DA, Chekhonin VP. Role of microRNAs in mechanisms of glioblastoma resistance to radio- and chemotherapy. Biochemistry (Mosc) 2013;78:325-34. [Crossref] [PubMed]
- Hochberg FH, Atai NA, Gonda D, et al. Glioma diagnostics and biomarkers: an ongoing challenge in the field of medicine and science. Expert Rev Mol Diagn 2014;14:439-52. [Crossref] [PubMed]
- Li M, Deng H, Peng H, et al. Functional nanoparticles in targeting glioma diagnosis and therapies. J Nanosci Nanotechnol 2014;14:415-32. [Crossref] [PubMed]
- Marvaso G, Barone A, Amodio N, et al. The current status of novel PET radio-pharmaceuticals in radiotherapy treatment planning of glioma. Curr Pharm Biotechnol 2014;14:1099-104. [Crossref] [PubMed]
- Giordano FA, Abo-Madyan Y, Brehmer S, et al. Intraoperative radiotherapy (IORT)—a resurrected option for treating glioblastoma? Transl Cancer Res 2014;3:94-105.
- Khasraw M, Ameratunga M, Grommes C. Bevacizumab for the treatment of high-grade glioma: an update after phase III trials. Expert Opin Biol Ther 2014;14:729-40. [Crossref] [PubMed]
- Thaci B, Brown CE, Binello E, et al. Significance of interleukin-13 receptor alpha 2-targeted glioblastoma therapy. Neuro Oncol 2014;16:1304-12. [Crossref] [PubMed]
- Prasanna A, Ahmed MM, Mohiuddin M, et al. Exploiting sensitization windows of opportunity in hyper and hypo-fractionated radiation therapy. J Thorac Dis 2014;6:287-302. [PubMed]
- Zhao B, Bian EB, Li J, et al. New advances of microRNAs in glioma stem cells, with special emphasis on aberrant methylation of microRNAs. J Cell Physiol 2014;229:1141-7. [Crossref] [PubMed]
- Palumbo S, Miracco C, Pirtoli L, et al. Emerging roles of microRNA in modulating cell-death processes in malignant glioma. J Cell Physiol 2014;229:277-86. [Crossref] [PubMed]
- Santovito D, Mezzetti A, Cipollone F. MicroRNAs and atherosclerosis: new actors for an old movie. Nutr Metab Cardiovasc Dis 2012;22:937-43. [Crossref] [PubMed]
- Besse A, Sana J, Fadrus P, et al. MicroRNAs involved in chemo- and radioresistance of high-grade gliomas. Tumour Biol 2013;34:1969-78. [Crossref] [PubMed]
- Ballman KV. Biomarker-based trials in neuro-oncology. Chin Clin Oncol 2015;4:38. [PubMed]
- Guz M, Rivero-Müller A, Okoń E, et al. MicroRNAs-role in lung cancer. Dis Markers 2014;2014:218169.
- Liao JM, Cao B, Zhou X, et al. New insights into p53 functions through its target microRNAs. J Mol Cell Biol 2014;6:206-13. [Crossref] [PubMed]
- Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays 2014;36:617-26. [Crossref] [PubMed]
- Ciafrè SA, Galardi S. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol 2013;10:935-42. [Crossref] [PubMed]
- Luo X, Stock C, Burwinkel B, et al. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One 2013;8:e62880 [Crossref] [PubMed]
- He H, Ni J, Huang J. Molecular mechanisms of chemoresistance in osteosarcoma Oncol Lett 2014;7:1352-62. (Review). [PubMed]
- Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics 2014;30:2243-6. [Crossref] [PubMed]
- Li Q, Wang JX, He YQ, et al. MicroRNA-185 regulates chemotherapeutic sensitivity in gastric cancer by targeting apoptosis repressor with caspase recruitment domain. Cell Death Dis 2014;5:e1197 [Crossref] [PubMed]
- Chen Z. Genomic analysis of drug resistant gastric cancer cell lines by combining mRNA and microRNA expression profiling. Cancer Lett 2014;350:43-51. [Crossref] [PubMed]
- Li X, Yang H, Tian Q, et al. Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. Neoplasma 2014;61:453-60. [Crossref] [PubMed]
- Jin HY, Lai M, Xiao C. microRNA-17~92 is a powerful cancer driver and a therapeutic target. Cell Cycle 2014;13:495-6. [Crossref] [PubMed]
- Garg N, Po A, Miele E, et al. microRNA-17-92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J 2013;32:2819-32. [Crossref] [PubMed]
- Jin HY, Oda H, Lai M, et al. MicroRNA-17~92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J 2013;32:2377-91. [Crossref] [PubMed]
- Shen Y, Lu L, Xu J, et al. Bortezomib induces apoptosis of endometrial cancer cells through microRNA-17-5p by targeting p21. Cell Biol Int 2013;37:1114-21. [Crossref] [PubMed]
- Khan AA, Penny LA, Yuzefpolskiy Y, et al. MicroRNA-17~92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood 2013;121:4473-83. [Crossref] [PubMed]
- Guo L, Xu J, Qi J, et al. MicroRNA-17-92a upregulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. J Cell Sci 2013;126:978-88. [Crossref] [PubMed]
- Zeller C, Hinzmann B, Seitz S, et al. SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene 2003;22:2972-83. [Crossref] [PubMed]
- Rimkus C, Martini M, Friederichs J, et al. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br J Cancer 2006;95:1419-23. [Crossref] [PubMed]
- Yang L, Liu M, Gu Z, et al. Overexpression of SASH1 related to the decreased invasion ability of human glioma U251 cells. Tumour Biol 2012;33:2255-63. [Crossref] [PubMed]
- Shah AH, Snelling B, Bregy A, et al. Discriminating radiation necrosis from tumor progression in gliomas: a systematic review what is the best imaging modality? J Neurooncol 2013;112:141-52. [Crossref] [PubMed]
- Hirst TC, Vesterinen HM, Sena ES, et al. Systematic review and meta-analysis of temozolomide in animal models of glioma: was clinical efficacy predicted? Br J Cancer 2013;108:64-71. [Crossref] [PubMed]
- Moore G, Collins A, Brand C, et al. Palliative and supportive care needs of patients with high-grade glioma and their carers: a systematic review of qualitative literature. Patient Educ Couns 2013;91:141-53. [Crossref] [PubMed]
- Sciumè G, Santoni A, Bernardini G. Chemokines and glioma: invasion and more. J Neuroimmunol 2010;224:8-12. [Crossref] [PubMed]
- Sherman JH, Hoes K, Marcus J, et al. Neurosurgery for brain tumors: update on recent technical advances. Curr Neurol Neurosci Rep 2011;11:313-9. [Crossref] [PubMed]
- Hemmat M, Rumple MJ, Mahon LW, et al. Short stature, digit anomalies and dysmorphic facial features are associated with the duplication of miR-17~92 cluster. Mol Cytogenet 2014;7:27. [Crossref] [PubMed]
- Li M, Guan X, Sun Y, et al. miR-92a family and their target genes in tumorigenesis and metastasis. Exp Cell Res 2014;323:1-6. [Crossref] [PubMed]
- Aviv T, Lin Z, Lau S, et al. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 2003;10:614-21. [Crossref] [PubMed]
- Koch CA, Anderson D, Moran MF, et al. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science 1991;252:668-74. [Crossref] [PubMed]
- Claudio JO, Zhu YX, Benn SJ, et al. HACS1 encodes a novel SH3-SAM adaptor protein differentially expressed in normal and malignant hematopoietic cells. Oncogene 2001;20:5373-7. [Crossref] [PubMed]
- Kim CA, Gingery M, Pilpa RM, et al. The SAM domain of polyhomeotic forms a helical polymer. Nat Struct Biol 2002;9:453-7. [PubMed]
- Kuribayashi K, Nakamura K, Tanaka M, et al. Essential role of protein kinase C zeta in transducing a motility signal induced by superoxide and a chemotactic peptide, fMLP. J Cell Biol 2007;176:1049-60. [Crossref] [PubMed]
- Martini M, Gnann A, Scheikl D, et al. The candidate tumor suppressor SASH1 interacts with the actin cytoskeleton and stimulates cell-matrix adhesion. Int J Biochem Cell Biol 2011;43:1630-40. [Crossref] [PubMed]
- Lin S, Zhang J, Xu J, et al. Effects of SASH1 on melanoma cell proliferation and apoptosis in vitro. Mol Med Rep 2012;6:1243-8. [PubMed]
- Sheyu L, Hui L, Junyu Z, et al. Promoter methylation assay of SASH1 gene in breast cancer. J BUON 2013;18:891-8. [PubMed]
- Zhou D, Wei Z, Deng S, et al. SASH1 regulates melanocyte transepithelial migration through a novel Gαs-SASH1-IQGAP1-E-Cadherin dependent pathway. Cell Signal 2013;25:1526-38. [Crossref] [PubMed]
- Liang F, Zhang S, Wang B, et al. Overexpression of integrin-linked kinase (ILK) promotes glioma cell invasion and migration and down-regulates E-cadherin via the NF-κB pathway. J Mol Histol 2014;45:141-51. [Crossref] [PubMed]
- Carraro G, El-Hashash A, Guidolin D, et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol 2009;333:238-50. [Crossref] [PubMed]
- Li SH, Guo J, Wu J, et al. miR-17 targets tissue inhibitor of metalloproteinase 1 and 2 to modulate cardiac matrix remodeling. FASEB J 2013;27:4254-65. [Crossref] [PubMed]
- Dakhlallah D, Batte K, Wang Y, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 2013;187:397-405. [Crossref] [PubMed]


