
Construction of diagnostic and prognostic models based on gene signatures of nasopharyngeal carcinoma by machine learning methods
Highlight box
Key findings
• Using the random forest algorithm, this work identified important gene signatures of nasopharyngeal carcinoma and constructed a diagnostic model using deep learning algorithms. The Lasso-Cox algorithm was used to create a prognostic prediction model, identifying potential therapeutic targets and pathways.
What is known and what is new?
• Some of the key gene signatures identified by previous studies through protein interaction networks are known;
• In this manuscript, we used the random forest algorithm to filter out new and more characteristic gene signatures and constructed the first diagnostic model for nasopharyngeal carcinoma using artificial neural networks. Prognostic models were also constructed to evaluate patient survival and identify some noteworthy therapeutic targets and pathways that are expected to inform treatment decisions.
What is the implication, and what should change now?
• Further studies are needed to investigate the mechanisms of these gene signatures.
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most prevalent cancers and has an uneven geographical distribution. The disease is particularly prevalent in East and Southeast Asia. The incidence of NPC in East Asia is extremely high, with an annual incidence of approximately 30 cases per 100,000 people (1), compared with less than 1 case per 100,000 people in the United States and Europe (2). Due to the hidden anatomical location and obscure symptoms, most patients cannot be diagnosed early. A study has reported that 70–80% of patients are already at a locally advanced stage when they are diagnosed (3), and this poses a serious threat to the life and health of the patients. Therefore, searching for gene signatures is needed for early diagnosis, survival prediction, and recurrence monitoring, optimizing medical interventions in NPC. It is of great significance to improve the quality of life and prolong the survival time of patient.
In recent years, the development of microarray technology and the development and wide application of RNA sequencing technology have facilitated the identification of various disease-associated gene signatures, which has greatly facilitated the research related to gene signatures and mechanisms, and provided a solid foundation for the establishment of diagnostic models related to NPC gene signatures (4). Currently, Chen et al. (5) have screened seven key NPC genes by constructing protein interaction networks through gene differential expression analysis, and Liu et al. (6) have also screened key genes and signaling pathways by the same method, which has contributed to the diagnosis of NPC.
Although the identified gene signatures can be used as diagnostic and predictive tools for NPC, due to the complex structure of NPC gene signatures, traditional diagnostic prediction models are not effective enough for NPC screening and early detection (7). Previous studies mostly used conventional logistic regression algorithms to construct diagnostic models, but their decision surfaces are linear and cannot be used to solve non-linear problems, with the disadvantage of being easily underfitted and less accurate, not being able to address multiple types of features or variables well (8,9). The main difficulty faced in building diagnostic models using gene expression data is how to find the meaningful classification features (10). The random forest (RF) algorithm has shown good performance in processing high-dimensional data, generalizing well, and maintaining accuracy for a large number of missing features (11). RF has a significant advantage over logistic regression algorithms for filtering features (12). The artificial neural network (ANN) is a deep learning algorithm that can be optimized by iteratively adjusting the weights of the connected neurons until global optimization is achieved, which is a significant advantage in the training of binary classification models (13). ANN have been used to construct breast cancer diagnostic models and in the imaging of lymph node metastases (14,15). In the field of NPC, ANN have been used to construct prediction models for radiotherapy complications of NPC (16). These previous studies provide a valuable reference for the application of ANN.
Most previous studies have used only a limited number of gene signatures to construct prognostic prediction models, and most of them have focused on gene signatures that affect overall survival (OS) (17-19). The efficacy of predictive models for OS and disease-free survival (DFS) based on multiple gene signatures have not been investigated. The effect of DFS-related gene signatures on prognosis has also not been discussed. Further studies are also needed to explore the mutations in prognosis-related gene signatures.
Therefore, in this study, an NPC diagnostic model was constructed based on the advantages of RF and ANN algorithms. The least absolute shrinkage and selection operator (Lasso) and Cox regression algorithm constructed prognostic models for OS and DFS. Finally, the mutation status of gene signatures that were significantly associated with prognosis was analyzed. It contributes to the early screening and treatment of NPC, and the study of its molecular mechanisms. The goal is to improve the quality of life and prolong the survival time of patients. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2700/rc).
Methods
Data acquisition and planning
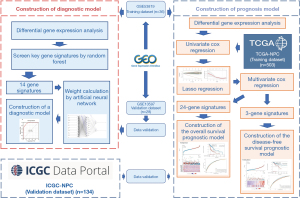
Microarray datasets GSE53819 (18 patients, 18 controls) and GSE13597 (15 patients, 13 controls) containing NPC patients and controls from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/; accessed 27 October 2022) were selected for the construction and validation of diagnostic models. GSE53819 and GSE13597 are the microarray dataset based on the GPL6480 platform (Agilent-014850 Whole Human Genome Microarray 4x44K G4112F). GSE53819 was used for differentially expressed genes (DEGs) screening and RF for feature screening and as a training set to calculate gene weights and develop diagnostic model using ANN. Then, we used the independent dataset GSE13597 for external validation of our model. RNA-sequencing (RNA-Seq) expression (level 3) profiles and corresponding clinical information for NPC were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.com/; accessed 2 February 2023) and International Cancer Genome Consortium (ICGC) database (https://icgc.org/; accessed 2 February 2023). Converting counts data to transcripts per million (TPM) and normalizing the data, keeping samples with clinical information at the same time. Finally, there are 503 samples in TCGA and 134 samples in ICGA were then utilized for further analysis. The TCGA dataset is the training set for the construction and training of the prognostic model, and the ICGC dataset is used for the validation of the prognostic model (Figure 1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of DEGs
The GSE53819 raw data file of the GEO database was downloaded using the GEOQuery package (version 2.54.1) in R statistical language (version 4.2.1). Remove the probes of multiple molecules corresponding to one probe. When the probe corresponding to the same molecule encountered, only the probe with the largest signal value will be saved. Next, we normalized the data using the NormalizeBetweenArrays function of the limma package (version 3.42.2) in R and analyzed the DEGs between the two groups, and the genes that met threshold |log2[fold change (FC)]| >2, false discovery rate (FDR) <0.01 and P.adj <0.05 were considered as highly valid DEGs (20,21).
Screening for key genes by random forest
The randomForest package (version 4.7) in R was used to construct the RF model for screening DEGs. In the random forest screener, the number of decision trees was initially set to 500 respectively. the error rate of each of the 1–500 decision trees was calculated and the optimal number of trees was determined by the number of trees with the best stability and the lowest error rate, which represents the constructed model with higher accuracy and small and stable model error (22). Subsequently, based on the selected parameters, the Gini coefficient method was used to obtain the dimensional importance values of all variables from the constructed random forest model. The DEGs with importance values greater than 1 were screened as significant genes for NPC and used for subsequent model construction and validation.
Construction of diagnostic models by ANNs
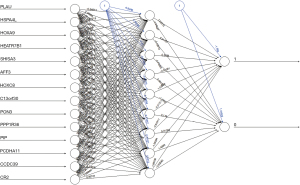
Based on the previous screening of DEGs, the expression data of DEGs was transformed into a “gene score” based on their expression levels, and normalized the data using the min-max method. For example, if the expression value of a down-regulated gene in a sample is higher than the median expression value of that gene in all samples, the expression value is 0, otherwise it is 1. Similarly, if the expression value of an up-regulated gene is higher, its expression is 1, otherwise it is 0. The neuralnet package (version 4.2) in R was used for ANN model construction, which has input, hidden, and output layers, and the number of hidden neuron layers should be two-thirds of the number of input layers. Based on the previous DEGs of 14, we set the number of hidden layers to 9 and the output layer to 2 nodes (NPC/Normal). Then, the formula of NeuralNPC was constructed to evaluate the classification score of NPC by multiplying the weight score by the expression level of each gene to calculate the classification score (23,24). Finally, the pROC package (version 1.17.0.1) in R was used to calculate the accuracy of the diagnostic prediction performance of the models constructed in this study (25).
Validation of the diagnostic model by area under the curve
The external dataset GSE13597 was used as a validation set for external validation of the accuracy of the constructed ANN model for NPC diagnosis. The R package pROC was used to plot the receiver operating characteristic (ROC) curves for each dataset and its diagnostic performance was evaluated by areas under the curve (AUC).
Construction of a prognostic model based on gene signatures
Univariate Cox regression analysis was used to evaluate the association between genes and NPC prognosis. DEGs that were significantly associated with NPC (P<0.05) were considered as candidate genes (26). Subsequently, the glmnet package in R was used to filter and remove genes that might result in overfitting the model by the Lasso algorithm. Finally, multivariate Cox analysis was applied to identify the optimal prognosis-related genes for the model. Risk scores were calculated based on a linear combination of Cox coefficients and gene expression. The following formulate was used for the analysis (27):
The median was used as a cut-off value to divide all NPC patients in the training set into two groups: high-risk group and a low-risk group. A high-risk score indicates that a low survival rate for NPC patients. Survival analysis was performed using the Survival package and the survminer package. ROC analysis for OS and DFS was used to evaluate the accuracy of the prognostic model. The survivalROC package was used to perform ROC analysis and AUC >0.60 was considered a valid predictive value. Risk score distribution plots between low- and high-risk groups, survival status scatter plots, and heat maps were also used to evaluate the model. Log-rank analysis was used to compare the predictive accuracy of the model and risk score (28).
Validation of the prognostic model
A sample of 134 patients from the ICGC database was used to validate the accuracy of the prognostic risk model. Survival analysis and time-dependent ROC analysis were used to validate the model. Risk score distribution plots, survival status scatter plots and heat maps were also used to evaluate the model.
Prognostic gene signature mutation
To further explore the value of prognostic gene signatures, the cBioPortal online website (http://www.cbioportal.org/; accessed 4 February 2023) was used to analyze the mutation profile of prognostic gene signatures in patients (29).
DEG enrichment analysis
Enrichment analysis of the DEGs was conducted by using GSEA software (version 3.0) from the GSEA (http://software.broadinstitute.org/gsea/index.jsp; accessed 5 February 2023). Samples were divided into high (≥50%) and low (<50%) expression groups based on their expression levels. The c2.cp.v7.2.symbols.gmt (Curated) subset from molecular signature dataset (http://www.gsea-msigdb.org/gsea/downloads.jsp; accessed 5 February 2023) was downloaded to analyze relevant pathways and molecular mechanisms. Based on gene expression profiles and phenotypic groupings, the minimum gene set was set at 5 and the maximum gene set was set at 800, with 1,000 resampling performed. Statistical significance was considered when the P value was <0.05 and the FDR was <0.25 (30).
Statistical analysis
The random forest algorithm was used for gene signatures screening, the ANN algorithm was used for model construction, and the absolute partial derivative of the error function <0.01 terminated the training. DFS and OS were calculated using Kaplan-Meier curves, and statistical differences were determined by log-rank test. The effects of various variables on DFS and OS were assessed by univariate and multivariate Cox regression models. Hazard ratio (HR) and 95% confidence interval (CI) were generated using Cox models. ROC analysis was performed to compare the predictive accuracy of gene signatures. P<0.05 was set as a statistically significant difference.
Results
Identification of the DEGs in the NPC
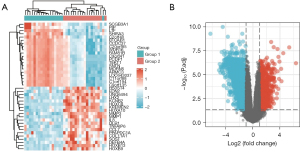
Based on previously set thresholds, a total of 582 DEGs were identified, including 156 upregulated DEGs and 426 downregulated DEGs, in 18 patients and 18 controls in the GSE53819 dataset. Using the limma package and visualized them with the ggplot2 (version 3.3.3) package and the ComplexHeatmap (version 2.2.0) package. Volcano maps and heat maps were plotted (Figure 2). These 582 DEGs with significant characteristics were applied as candidate genes in the subsequent analysis.
Random forest screens key genes associated with NPC
The 582 DEGs were applied to the random forest model, and the optimal number of trees (n=27) was determined by calculating the error rate for each of the 1–500 trees (Figure 3A). The Gini coefficient method measures the importance of all variables according to the reduction in mean square error and model accuracy, and the 15 important gene signatures were output in (Figure 3B). Finally, the 14 DEGs with MeanDecreaseGini >1 (PLAU, HSPA4L, HOXA9, HEATR7B1, SHISA3, AFF3, HOXC8, C13orf30, PON3, PPP1R36, PIP, PCDHA11, CCDC39, CR2) was selected as important gene signatures for subsequent analysis. We visualized these 14 DEGs in the samples and plotted the expression heat map in the samples (Figure 3C).

Construction and validation of NPC diagnostic models based on ANNs
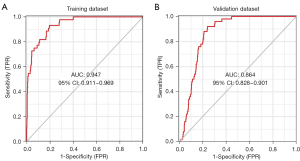
The first step was data preprocessing of the normalized data, converting the expression of previously screened key genes into gene scores of 0 or 1. The analysis was performed using an ANN to calculate optimized weights for all genes. Then, an ANN diagnostic model containing 14 input layers, 9 hidden layers and two output layers was constructed and the ANN was visualized (Figure 4). The entire training was performed in 6,782 steps with a termination condition of absolute partial derivative of the error function <0.01. After the training set model was built, we plotted the AUC for evaluating the classification performance of the model. The model had an AUC of 0.947 (95% CI: 0.911–0.969) in the training dataset (Figure 5A) and an AUC of 0.864 (95% CI: 0.828–0.901) in the validation dataset (Figure 5B), indicating that the ANN model has a good performance in diagnosing NPC. The above results indicated that a diagnostic prediction model for NPC was successfully constructed using differential gene expression between NPC and normal samples.
Prognostic gene signatures model construction
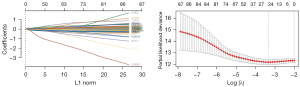
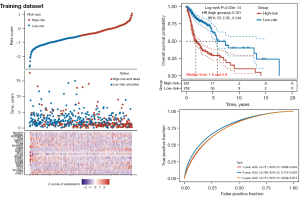
Predictive models for DFS and OS were constructed in this study. The OS prediction model included 637 patients, which were divided into a training set (n=503) and a validation set (n=134) based on different sources of data sets from different centers. The DFS prediction model included 289 patients divided into training set (n=230) and validation set (n=59) for construction and validation, respectively. The univariate Cox analysis screened 87 gene signatures, followed by the selection of the optimal parameter, “lambda.min”, through Lasso regression with 1,000 iterations. Twenty-four gene signatures were found to be significantly associated with OS by Lasso regression, as shown in (Figure 6). To further filter these 24-gene signatures, 3 gene signatures were found to be significantly associated with DFS by multivariate Cox. Based on the previous risk score formula, patients were divided into a high-risk group (OS: n=251; DFS: n=115) and a low-risk group (OS: n=252; DFS: n=115). Time-dependent ROC and Kaplan-Meier curves were used to evaluate the predictive potential of patients in the training set at 1-, 3-, and 5-year OS (Figure 7) and DFS (Figure 8). The AUC shows the predictive effect of our model in the training set
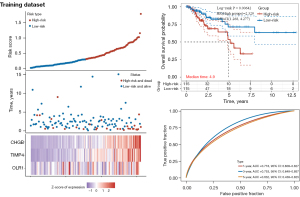
Validation of the prognosis prediction model based on gene signatures
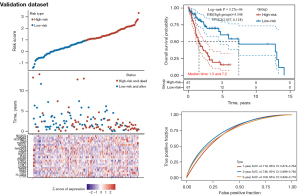
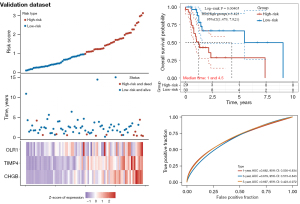
The validation set data was also evaluated using the calculated risk score, which divided patients into high-risk (OS: n=67; DFS: n=29) and low-risk (OS: n=67; DFS: n=30) groups. The predictive ability of the prediction models was evaluated using time-dependent ROC and Kaplan-Meier curves for 1, 3, and 5 years of survival in the validation set for both OS and DFS (Figures 9,10). The results showed that the high-risk group had shorter survival times compared to the low-risk group, and the prediction model had a robust predictive effect.
Mutation characteristics and gene set enrichment analysis
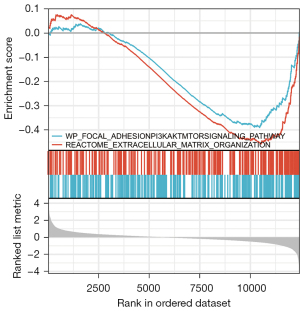
cBioPortal was used to explore the mutational profile of the 24-gene signatures (Figure 11). Amplification of SHANK2 was observed in 25% of the samples, while both amplification and deep deletion of STAR were found in 8% of the samples. MOGAT2 and CCBE1 were found to have amplification and deep deletion in 4% of samples. The other genes showed no significant mutational changes in the sample. GSEA enrichment analysis showed that genes in the high-risk and low-risk groups were enriched in the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway and extracellular matrix organization (Figure 12).
Discussion
NPC is a prevalent malignancy in East and Southeast Asia, and patients with early detection often have a good prognosis (31). Therefore, early prediction and diagnosis of NPC can inform and improve the success rate of early interventions for the treatment of NPC. Considering the lack of effective methods for early screening and diagnosis of NPC due to the lack of obvious early symptoms, as well as the lack of characteristic disease markers that can be used in clinical practice, it is important to develop predictive models for early diagnosis and screening and prognosis of NPC based on gene signatures (32).
With the development of various sequencing technologies as well as public databases, it is now possible to use publicly available disease data for studies related to the diagnostic identification and prognostic gene signature of diseases. Advances in computer science and technology have enabled the use of methods such as machine learning and deep learning applied to the study of bioinformatics and imaging analysis, a technical field that shows great potential (33-35). In the present study, we further explore diagnostic NPC gene signature at the molecular level, and machine learning approaches have shown great advantages in key gene selection and classification, which have been previously studied using random forest algorithms and ANNs in areas such as heart failure (36).
The problem we face is the binary classification problem of whether a patient has NPC or not, and deep learning is the most widely used machine learning method in medical research, providing a good processing model for developing complex, automatic and objective algorithms for analyzing high-dimensional and multimodal biomedical data (37). RF has a very high predictive accuracy and can provide information about the importance of variables for classification (38). ANN differs from traditional regression analysis in that neural networks can analyze nonlinear data due to their data processing capabilities. With the selection of appropriate input and output layers, functional relationships with infinitely close correlations between the input and output layers can be discovered by learning and debugging large amounts of clinical data through network models (39). The network model is trained by providing the neural network with input and output layers and the connection weights can be adjusted during iterations to match the output with the actual output until the desired result is obtained (40). Based on these advantages, we chose a random forest combined with an ANN approach to construct a diagnostic prediction model for NPC. The prognostic survival information was a continuous classification problem and required multiple hidden layers in ANN, but using too many hidden layers would result in a large computational burden and negatively affect the accuracy of the model (41). Therefore, ANN was not selected to build the prediction model. The Lasso-Cox regression algorithm has been applied in continuous variable classification problems and has achieved satisfactory results in the construction of prognostic models. Therefore, Lasso-Cox regression was used to construct prognostic prediction models based on risk scores (42).
In this study, DEGs associated with NPC were identified by differential gene expression analysis. subsequently, 14 key DEGs were identified by the RF algorithm, and a diagnostic model for NPC was developed by the ANN model. Among our screened DEGs, we found that PLAU, HOXA9, HSPA4L, and HOXC8 were highly expressed in NPC patient samples and lowly expressed in normal control samples. PPP1R36, PIP, PCDHA11, CCDC39, CR2, C13orf30, PON3, HEATR7B1, SHISA3 and AFF3, on the other hand, were lowly expressed in NPC patient samples and highly expressed in control samples (Figure 3C).
PLAU has been shown that its high expression can promote cell proliferation and epithelial mesenchymal transition in head and neck squamous cell carcinoma by regulating cell-matrix adhesion, tissue migration and extracellular matrix organization, and overexpression often has a poor prognosis (43). The potent oncogene miR-497 can inhibit the cancer phenotype of NPC by targeting HSPA4L, which can inhibit the proliferation migration and induce apoptosis of NPC cells, and has potential targeted therapeutic implications (44). HOXA9 was found to have its overexpression correlated with NPC tumor development differentiation and prognosis, and controlled studies have now found that patients with high HOXA9 expression progress from nasopharyngitis to HOXC8 has been found to regulate glycolysis and regulate the expression of genes related to the tricarboxylic acid cycle and NPC (45,46). CCDC39 has been found to improve the prognosis of NPC patients by regulating immune cell mechanisms, but causes low expression of memory B cells which does not contribute to patient prognosis (47). Inflammatory and immune responses during Epstein-Barr virus (EBV) infection contribute to the development of NPC, and a 5'-untranslated region (UTR) functional polymorphism of CR2 was found to be associated with EBV susceptibility (48). SHISA3 was found to promote NPC metastasis by decreasing the stability of SGSM1, and to cause this gene to be patients and SHISA3 was found to inhibit NPC invasion and metastasis (49).
Our study further validated the value of these key genes in NPC, and found that PPP1R36, PIP, PCDHA1, C13orf30, PON3, HEATR7B1, and AFF3 were found to have different roles in the mechanism, diagnosis, and prognosis of other cancers (50), but relevant studies in the field of NPC have not been conducted yet, which provides a direction for further research.
In the prognostic model, we were surprised to find that PLAU was significantly associated with both the diagnosis and prognosis of NPC. Dong et al. (51) have demonstrated that knockdown of MKLK can inhibit cell proliferation and epithelial mesenchymal transition caused by PLAU and reduce the invasion and metastasis of treatment-induced radioresistant NPC cells. Indicated that PLAU is a potentially important target for improving patient prognosis. Three gene signatures were used to construct a DFS prediction model, with the results showing that they were down-regulated in the low-risk group and up-regulated in the high-risk group. High expression of these 3 genes may indicate a poor prognosis. Liu et al. (52) analyzed the RNA-Seq from peripheral blood mononuclear cells of NPC and revealed that ORL1 was significantly associated with radiation response and had a negative impact on prognosis in patients with high expression levels. Additionally, another study found that there was a positive numerical correlation between PD-L1 and OLR1. The positive numerical correlation between OLR1 and CD8+ may further supports the potential use of OLR1 as a biomarker for immunotherapy (53). TIMP4 has yet to be studied in the context in NPC. However, it has been found to be involved in several processes in other cancers, including cell invasion and migration, cell proliferation and apoptosis, and angiogenesis (54). TIMP4 with high expression has been linked to poor prognosis in two studies of head and neck squamous cell carcinomas (55). Zhou et al. (56) discovered that CHGB promotes the development of NPC and is associated with advanced lymph node metastasis. Furthermore, CHGB was also identified as a poor prognostic significant genetic signature in a study.
In this study, 4 prognostic gene signatures were found to have more significant mutations in the sample. SHANK2 is a commonly amplified gene in human cancers. A knockdown of SHANK2 restores Hippo signaling in cancer cells, resulting in reduced cell proliferation (57). Xu et al. (58) found that no cancer cell mass was found in vivo after SHANK2 depletion. These findings further validate the role of SHANK2 in the Hippo signaling pathway and indicate that SHANK2 may be a promising target to improve patient prognosis and treatment. MOGAT2, a gene signature significantly associated with metabolism, was found to have a negative impact on prognosis in patients with hepatocellular carcinoma, and Blanc et al. (59) reported that MOGAT2 could be applied as a novel biomarker affecting the treatment of colorectal cancer metastases. CCBE1 is a noteworthy gene that is upregulated upon overexpression of FGF14, and when downregulated, it is linked to reduced OS and DFS in patients (60). The gene exhibits a deep deletion when downregulated in different cancer patients and is associated with poor prognosis. Our study confirms that this situation is also present in some NPC patients. Moreover, our findings provide further support for the potential therapeutic utility of inhibitors targeting FGF14 in patients with NPC who exhibit CCBE1 downregulation. These newly established inhibitors hold promise as a promising therapeutic approach for these patients. STAR protein is an integral component in the regulation of cholesterol transport and steroid hormone biosynthesis. The role of the STAR protein in these processes has been well established in the literature (61). In our recent study, we observed significant mutations in the STAR gene. Manna et al. (62) have reported elevated expression levels of STAR in hormone-responsive breast cancer cells, and this upregulation has been linked to the growth and migration of these cells. Their findings suggest that amplification of the STAR gene may contribute to the decreased survival rates of patients diagnosed with this disease. These results highlight the importance of further investigating the potential of the STAR gene as a therapeutic target in the treatment of NPC.
GSEA analysis revealed that genes associated with prognosis of NPC were mainly enriched in two pathways, PI3K/Akt and extracellular matrix organization. PI3K/Akt is a classical pathway. Chen et al. (63) found that mutations in the PI3K/Akt/mTOR/AMPK signaling pathway are associated with a poor prognosis in patients with NPC. In another study, YBX3 was found to mediate the metastasis of NPC via the PI3K/Akt signaling pathway, and YBX3 was upregulated in various tumor cells, and this upregulation was associated with tumor cell proliferation and chemotherapy resistance (64). Xie et al. (65) also found that C2orf40 inhibited NPC cell metastasis and regulated chemoresistance and radioresistance by affecting the cell cycle and activating the PI3K/Akt/mTOR signaling pathway. Therefore, this signaling pathway is a potential therapeutic direction for NPC research. In several studies, extracellular matrix organization has been identified as one of the key pathways in NPC. Surprisingly, the PLAU, which was significantly associated with both diagnosis and prognosis in this study, promoted cancer metastasis and proliferation through the extracellular matrix pathway. Therefore, further researches on the mechanism of PLAU in NPC are needed, which may be a new target to improve the treatment and prognosis of NPC.
In this study, the diagnostic model based on machine learning and deep learning algorithms can effectively screen the occurrence of NPC at an early stage. It helps to provide a foundation for early intervention treatment of patients with NPC, making sure treatments such as radiotherapy are giving at the optimal stage. Additionally, patients were grouped according to risk scores, and a prognostic model was constructed to predict OS and DFS in high-risk and low-risk patients. Some of these gene signatures used to construct the prognostic models were found to be potentially valuable, and some of them were found to be adjuvant to immunotherapy and radiotherapy, which could be potential therapeutic targets to improve patient prognosis. The OS and DFS prediction models based on patient risk scores and gene marker mutation analysis are also used to provide references for individualized treatment decisions such as surgery, chemotherapy or radiotherapy, as well as potential gene-targeted therapies possibility for patients with different risk levels.
There are limitations in this study. For several reasons, we did not perform in vivo or in vitro experiments to validate our diagnostic prediction model. However, the model we developed was validated with the current sample size to obtain robust performance and clinical value, so we will further investigate our model in a clinical setting.
Conclusions
In this study, machine learning methods were used to construct a diagnostic and a prognostic prediction model for NPC based on gene signatures. This model serves as a valuable reference for the early diagnosis and prognostic evaluation of the disease and its potential molecular mechanisms and therapeutic applications. However, further research is necessary to investigate the mechanisms and roles associated with the relevant gene signatures to verify the roles of these gene signatures in NPC.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2700/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2700/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer 2011;30:114-9. [Crossref] [PubMed]
- Lam WKJ, Chan JYK. Recent advances in the management of nasopharyngeal carcinoma. F1000Res 2018;7:F1000 Faculty Rev 1829;
- Chen YP, Chan ATC, Le QT, et al. Nasopharyngeal carcinoma. Lancet 2019;394:64-80. [Crossref] [PubMed]
- Huang X, Liu S, Wu L, et al. High Throughput Single Cell RNA Sequencing, Bioinformatics Analysis and Applications. Adv Exp Med Biol 2018;1068:33-43. [Crossref] [PubMed]
- Chen F, Shen C, Wang X, et al. Identification of genes and pathways in nasopharyngeal carcinoma by bioinformatics analysis. Oncotarget 2017;8:63738-49. [Crossref] [PubMed]
- Liu K, Kang M, Zhou Z, et al. Bioinformatics analysis identifies hub genes and pathways in nasopharyngeal carcinoma. Oncol Lett 2019;18:3637-45. [Crossref] [PubMed]
- Wang X, Zhang Y, Zhou P, et al. A supervised protein complex prediction method with network representation learning and gene ontology knowledge. BMC Bioinformatics 2022;23:300. [Crossref] [PubMed]
- Li G, Huang B, Wu H, et al. Development of novel gene signatures for the risk stratification of prognosis and diagnostic prediction of osteosarcoma patients using bioinformatics analysis. Transl Cancer Res 2022;11:2374-87. [Crossref] [PubMed]
- Westreich D, Lessler J, Funk MJ. Propensity score estimation: neural networks, support vector machines, decision trees (CART), and meta-classifiers as alternatives to logistic regression. J Clin Epidemiol 2010;63:826-33. [Crossref] [PubMed]
- Wu Y, Chen H, Li L, et al. Construction of Novel Gene Signature-Based Predictive Model for the Diagnosis of Acute Myocardial Infarction by Combining Random Forest With Artificial Neural Network. Front Cardiovasc Med 2022;9:876543. [Crossref] [PubMed]
- Kursa MB. Robustness of Random Forest-based gene selection methods. BMC Bioinformatics 2014;15:8. [Crossref] [PubMed]
- Grobman WA, Stamilio DM. Methods of clinical prediction. Am J Obstet Gynecol 2006;194:888-94. [Crossref] [PubMed]
- Camacho DM, Collins KM, Powers RK, et al. Next-Generation Machine Learning for Biological Networks. Cell 2018;173:1581-92. [Crossref] [PubMed]
- Sunny J, Rane N, Kanade R, et al. Breast cancer classification and prediction using machine learning. International Journal of Engineering Research and Technology 2020;9:576-80.
- Geng M, Geng M, Wei R, et al. Artificial intelligence neural network analysis and application of CT imaging features to predict lymph node metastasis in non-small cell lung cancer. J Thorac Dis 2022;14:4384-94. [Crossref] [PubMed]
- Wang T, Hu J, Huang Q, et al. Development of a normal tissue complication probability (NTCP) model using an artificial neural network for radiation-induced necrosis after carbon ion re-irradiation in locally recurrent nasopharyngeal carcinoma. Ann Transl Med 2022;10:1194. [Crossref] [PubMed]
- Zhou J, Zhang B, Zhang X, et al. Identification of a 3-miRNA Signature Associated With the Prediction of Prognosis in Nasopharyngeal Carcinoma. Front Oncol 2022;11:823603. [Crossref] [PubMed]
- Zhu GL, Yang KB, Xu C, et al. Development of a prediction model for radiotherapy response among patients with head and neck squamous cell carcinoma based on the tumor immune microenvironment and hypoxia signature. Cancer Med 2022;11:4673-87. [Crossref] [PubMed]
- Wang YQ, Zhang Y, Jiang W, et al. Development and validation of an immune checkpoint-based signature to predict prognosis in nasopharyngeal carcinoma using computational pathology analysis. J Immunother Cancer 2019;7:298. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Quist J, Taylor L, Staaf J, et al. Random Forest Modelling of High-Dimensional Mixed-Type Data for Breast Cancer Classification. Cancers (Basel) 2021;13:991. [Crossref] [PubMed]
- Beck MW. NeuralNetTools: Visualization and Analysis Tools for Neural Networks. J Stat Softw 2018;85:1-20. [Crossref] [PubMed]
- Bianconi A, Von Zuben CJ, Serapião AB, et al. Artificial neural networks: a novel approach to analysing the nutritional ecology of a blowfly species, Chrysomya megacephala. J Insect Sci 2010;10:58. [Crossref] [PubMed]
- Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [Crossref] [PubMed]
- Zhou L, Yu Y, Wen R, et al. Development and Validation of an 8-Gene Signature to Improve Survival Prediction of Colorectal Cancer. Front Oncol 2022;12:863094. [Crossref] [PubMed]
- Wang Z, Pan L, Guo D, et al. A novel five-gene signature predicts overall survival of patients with hepatocellular carcinoma. Cancer Med 2021;10:3808-21. [Crossref] [PubMed]
- Zhang Z, Lin E, Zhuang H, et al. Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma. Cancer Cell Int 2020;20:27. [Crossref] [PubMed]
- Unberath P, Mahlmeister L, Reimer N, et al. Searching of Clinical Trials Made Easier in cBioPortal Using Patients' Genetic and Clinical Profiles. Appl Clin Inform 2022;13:363-9. [Crossref] [PubMed]
- Fang Z, Liu X, Peltz G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 2023;39:btac757. [Crossref] [PubMed]
- Dionisi F, Croci S, Giacomelli I, et al. Clinical results of proton therapy reirradiation for recurrent nasopharyngeal carcinoma. Acta Oncol 2019;58:1238-45. [Crossref] [PubMed]
- Li H, Kong Z, Xiang Y, et al. The role of PET/CT in radiotherapy for nasopharyngeal carcinoma. Front Oncol 2022;12:1017758. [Crossref] [PubMed]
- Kriegeskorte N, Golan T. Neural network models and deep learning. Curr Biol 2019;29:R231-6. [Crossref] [PubMed]
- Auslander N, Gussow AB, Koonin EV. Incorporating Machine Learning into Established Bioinformatics Frameworks. Int J Mol Sci 2021;22:2903. [Crossref] [PubMed]
- Currie G, Hawk KE, Rohren E, et al. Machine Learning and Deep Learning in Medical Imaging: Intelligent Imaging. J Med Imaging Radiat Sci 2019;50:477-87. [Crossref] [PubMed]
- Tian Y, Yang J, Lan M, et al. Construction and analysis of a joint diagnosis model of random forest and artificial neural network for heart failure. Aging (Albany NY) 2020;12:26221-35. [Crossref] [PubMed]
- Savargiv M, Masoumi B, Keyvanpour MR. A New Random Forest Algorithm Based on Learning Automata. Comput Intell Neurosci 2021;2021:5572781. [Crossref] [PubMed]
- Anaissi A, Kennedy PJ, Goyal M, et al. A balanced iterative random forest for gene selection from microarray data. BMC Bioinformatics 2013;14:261. [Crossref] [PubMed]
- Koulaouzidis G, Jadczyk T, Iakovidis DK, et al. Artificial Intelligence in Cardiology-A Narrative Review of Current Status. J Clin Med 2022;11:3910. [Crossref] [PubMed]
- Alaskar H, Hussain A, Almaslukh B, et al. Deep Learning Approaches for Automatic Localization in Medical Images. Comput Intell Neurosci 2022;2022:6347307. [Crossref] [PubMed]
- Tu JV. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J Clin Epidemiol 1996;49:1225-31. [Crossref] [PubMed]
- He J, Li W, Li Y, et al. Construction of a prognostic model for lung adenocarcinoma based on bioinformatics analysis of metabolic genes. Transl Cancer Res 2020;9:3518-38. [Crossref] [PubMed]
- Chen G, Sun J, Xie M, et al. PLAU Promotes Cell Proliferation and Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. Front Genet 2021;12:651882. [Crossref] [PubMed]
- Wang S, Mo Y, Midorikawa K, et al. The potent tumor suppressor miR-497 inhibits cancer phenotypes in nasopharyngeal carcinoma by targeting ANLN and HSPA4L. Oncotarget 2015;6:35893-907. [Crossref] [PubMed]
- Liu T, Ji C, Sun Y, et al. HOXA9 Expression is Associated with Advanced Tumour Stage and Prognosis in Nasopharyngeal Carcinoma. Cancer Manag Res 2021;13:4147-54. [Crossref] [PubMed]
- Jiang Y, Yan B, Lai W, et al. Repression of Hox genes by LMP1 in nasopharyngeal carcinoma and modulation of glycolytic pathway genes by HoxC8. Oncogene 2015;34:6079-91. [Crossref] [PubMed]
- Yang Y, Zhang P, Zhang H, Lu J. Immunocyte infiltration characteristics of gene expression profile in nasopharyngeal carcinoma and clinical significance. Chinese Journal of Cellular and Molecular Immunology 2020;36:1069-75. [PubMed]
- Fan Q, He JF, Wang QR, et al. Functional polymorphism in the 5'-UTR of CR2 is associated with susceptibility to nasopharyngeal carcinoma. Oncol Rep 2013;30:11-6. [Crossref] [PubMed]
- Zhang J, Li YQ, Guo R, et al. Hypermethylation of SHISA3 Promotes Nasopharyngeal Carcinoma Metastasis by Reducing SGSM1 Stability. Cancer Res 2019;79:747-59. [Crossref] [PubMed]
- Schweikert EM, Devarajan A, Witte I, et al. PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ 2012;19:1549-60. [Crossref] [PubMed]
- Dong Y, Sun Y, Huang Y, et al. Depletion of MLKL inhibits invasion of radioresistant nasopharyngeal carcinoma cells by suppressing epithelial-mesenchymal transition. Ann Transl Med 2019;7:741. [Crossref] [PubMed]
- Liu G, Zeng X, Wu B, et al. RNA-Seq analysis of peripheral blood mononuclear cells reveals unique transcriptional signatures associated with radiotherapy response of nasopharyngeal carcinoma and prognosis of head and neck cancer. Cancer Biol Ther 2020;21:139-46. [Crossref] [PubMed]
- Liu B, Wang Z, Gu M, et al. GEO Data Mining Identifies OLR1 as a Potential Biomarker in NSCLC Immunotherapy. Front Oncol 2021;11:629333. [Crossref] [PubMed]
- Solga R, Behrens J, Ziemann A, et al. CRN2 binds to TIMP4 and MMP14 and promotes perivascular invasion of glioblastoma cells. Eur J Cell Biol 2019;98:151046. [Crossref] [PubMed]
- Zou M, Zhang C, Sun Y, et al. Comprehensive analysis of matrix metalloproteinases and their inhibitors in head and neck squamous cell carcinoma. Acta Oncol 2022;61:505-15. [Crossref] [PubMed]
- Zhou G, Zhai Y, Cui Y, et al. MDM2 promoter SNP309 is associated with risk of occurrence and advanced lymph node metastasis of nasopharyngeal carcinoma in Chinese population. Clin Cancer Res 2007;13:2627-33. [Crossref] [PubMed]
- Freier K, Sticht C, Hofele C, et al. Recurrent coamplification of cytoskeleton-associated genes EMS1 and SHANK2 with CCND1 in oral squamous cell carcinoma. Genes Chromosomes Cancer 2006;45:118-25. [Crossref] [PubMed]
- Xu L, Li P, Hao X, et al. SHANK2 is a frequently amplified oncogene with evolutionarily conserved roles in regulating Hippo signaling. Protein Cell 2021;12:174-93. [Crossref] [PubMed]
- Blanc V, Riordan JD, Soleymanjahi S, et al. Apobec1 complementation factor overexpression promotes hepatic steatosis, fibrosis, and hepatocellular cancer. J Clin Invest 2021;131:e138699. [Crossref] [PubMed]
- Turkowski K, Herzberg F, Günther S, et al. Fibroblast Growth Factor-14 Acts as Tumor Suppressor in Lung Adenocarcinomas. Cells 2020;9:1755. [Crossref] [PubMed]
- Manna PR, Ahmed AU, Molehin D, et al. Hormonal and Genetic Regulatory Events in Breast Cancer and Its Therapeutics: Importance of the Steroidogenic Acute Regulatory Protein. Biomedicines 2022;10:1313. [Crossref] [PubMed]
- Manna PR, Ahmed AU, Yang S, et al. Genomic Profiling of the Steroidogenic Acute Regulatory Protein in Breast Cancer: In Silico Assessments and a Mechanistic Perspective. Cancers (Basel) 2019;11:623. [Crossref] [PubMed]
- Chen Y, He Q, Ma H, et al. Relationship of PI3K-Akt/mTOR/AMPK signaling pathway genetic mutation with efficacy and prognosis in nasopharyngeal carcinoma. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2022;47:165-73. [PubMed]
- Fan X, Xie X, Yang M, et al. YBX3 Mediates the Metastasis of Nasopharyngeal Carcinoma via PI3K/AKT Signaling. Front Oncol 2021;11:617621. [Crossref] [PubMed]
- Xie Z, Li W, Ai J, et al. C2orf40 inhibits metastasis and regulates chemo-resistance and radio-resistance of nasopharyngeal carcinoma cells by influencing cell cycle and activating the PI3K/AKT/mTOR signaling pathway. J Transl Med 2022;20:264. [Crossref] [PubMed]



