
LINC00638/hsa-miR-29b-3p axis-mediated high expression of CDCA4 correlates with tumor immune infiltration and hepatocellular carcinoma progression
Highlight box
Key findings
• The low expression of cell division cycle-associated protein 4 (CDCA4) significantly improves the prognosis of liver hepatocellular carcinoma (LIHC) patients and may potentially be a new biomarker for LIHC prognosis prediction.
What is known and what is new?
• CDCA4 has been studied in tumors, such as lung squamous cell carcinoma, melanoma, osteosarcoma, and triple-negative breast cancer; however, no in-depth studies of CDCA4 in LIHC have been conducted.
• We examined the correlation between CDCA4 expression and its adverse clinical parameters. We also investigated the mechanism of CDCA4 regulation that may be related to the occurrence of LIHC, and the relationship between CDCA4 expression and immune infiltration.
What is the implication, and what should change now?
• Long non-coding RNA-LINC00638/hsa-miR-29b-3p/CDCA4 is a potential regulatory pathway in LIHC. Our findings suggest a new perspective for the development of LIHC anti-cancer strategies.
Introduction
Primary liver cancer has become the second most lethal cancer in the world, and it has the second highest fatality rate among digestive system malignancies in China (1). Liver hepatocellular carcinoma (LIHC) is the most prevalent primary liver cancer, and accounts for approximately 90% of all cases (2). LIHC has the characteristics of hidden onset, difficult early diagnosis, rapid progression, and high recurrence and metastasis rate. Its standard treatment methods include surgical resection, chemotherapy, radiation therapy, radiofrequency ablation, vascular embolization, or liver transplantation (3). Except for early small liver cancer resection surgery, there is still a lack of effective treatment methods for advanced LIHC (4). The prognosis of LIHC is poor, with a median survival time of approximately 11 months (5). Molecular marker screening remains a promising area of research that could extend understandings of the process of liver cancer. The capacity to investigate the gene expression profile of liver cancer has been substantially aided by the advent of high-throughput gene chip and sequencing technologies that make it possible to quickly screen genes and discover the mechanisms of action of key genes.
Cell division cycle-associated protein 4 (CDCA4) encodes a protein composed of 241 amino acids with a molecular weight of approximately 26 kDa (6). Its preferential expression in adult bone marrow hematopoietic progenitor cells led to its classification as a hematopoietic progenitor protein (HEPP) after its discovery by the differential screening of mouse hematopoietic stem cells using a reduced complementary DNA (cDNA) library. The function of HEPP is unknown; however, HEPP is missing in invertebrates, but is highly evolved and conserved in vertebrates, which suggests that HEPP may have a highly conserved unknown function (6). Walker et al. (7) conducted a guilty-by-association co-expression analysis on 1,176 human cDNA libraries to discover unknown human cell cycle genes, and found 8 previously unrecognized cell cycle genes [cell division cycle-associated protein1-8 (CDCA1-8)], which are highly co-expressed with many other known cell cycle genes, such as cyclin dependent kinase 1 (CDC2), cell division cycle 7 (CDC7), cell division cycle 23 (CDC23), and cyclin. This finding suggests that CDCA4 is involved in the regulation of the cell cycle (8,9). Studies have also been conducted on CDCA4 in tumors. For example, research has shown that microRNA-497-5p (miR-497-5p) inhibits the progression of lung squamous cell carcinoma by downregulating CDCA4 (10), the upregulation of microRNA-29c-3p (miR-29c-3p) prevents melanoma progression by inhibiting the expression of CDCA4 (11), the miR-503-5p/CDCA4 axis, which is mediated by Circ_0010220, contributes to the tumorigenesis of osteosarcoma progression (12), and signal transducer and activator of transcription 1 (STAT1) mediates the transcription of CircIFI30 and promotes the progression of triple-negative breast cancer by upregulating CDCA4 (13). However, CDCA4 in LIHC has not been well studied.
We sought to examine the correlation between CDCA4 expression and its clinical relevance using the Kaplan-Meier (KM) plotter using data from The Cancer Genome Atlas (TCGA), Genotype Tissue Expression (GTEX), Human Protein Atlas (HPA), and the University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN) databases. Additionally, the mechanism of CDCA4 regulation that may be implicated in the development of LIHC was investigated, as was the association between CDCA4 expression and immune infiltration. We conducted a number of studies, including correlation, expression, and survival analyses, to determine which non-coding RNAs (ncRNAs) are linked to increased CDCA4 expression in LIHC. We identified the LINC00638/hsa-miR-29b-3p axis as the most likely upstream ncRNA related pathway for CDCA4 in LIHC. In summary, our study confirms the potential role of CDCA4 in regulating tumor progression and its potential application in the diagnosis and prognosis evaluation of LIHC. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-569/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Analyses of data, including survival, correlation, and differential expression analyses
In the current study, the raw RNA-sequencing data and their clinical manifestation data were retrieved from 2 databases (i.e., the GTEX and TCGA databases). Data were acquired for 33 different LIHC cancers and healthy samples for the downstream analysis. We retrieved data from the liver cancer tissue and normal liver tissue through The Cancer Genome Atlas (TCGA). All analysis methods are performed using R software version v3.6.3 The “ggplot2”, “survminer’, and “survival” R programs (http://bioconductor.org/) were used to plot the expression analysis results and the KM survival curves. Significance was determined using the log-rank test, and univariate Cox proportional hazards regression was used to estimate P values and hazard ratios (HR) along with 95% confidence intervals (CI) in the KM curves. We used the “ggstatsplot” R (http://bioconductor.org/) tool to visualize the results of the 2-gene correlation analysis. A Pearson correlation or Spearman correlation analysis was used to evaluate the correlations between the quantitative data.
UALCAN database
To determine the protein-level expression of CDCA4 in LIHC, we used the UALCAN database (http://ualcan.path.uab.edu/), an online resource for interpreting TCGA gene expression data.
KM plotter analysis
The effect of several genes on the prognosis of 21 distinct cancer types was investigated using the online database KM plotter (http://kmplot.com/analysis/). We investigated the predictive significance of the microRNAs (miRNAs) in LIHC using the KM plotter.
PrognoScan database analysis
The PrognoScan database (http://www.abren.net/PrognoScan/). was used to analyze the correlation between the expression of CDCA4 and the total survival time [overall survival (OS)] in patients with LIHC.
HPA
The HPA database (https://proteinatlas.org/) contains protein-level gene expression profiling data from both normal and cancerous human tissues. In this study, the expression of CDCA4 in the LIHC tumor tissues was investigated.
ENCORI database analysis
The ENCORI database was used to determine the respective correlations among miRNA-ncRNA and miRNA-mRNA. Thus, we used ENCORI to predict which upstream miRNAs and long non-coding RNAs (lncRNAs) interact with CDCA4 and hsa-miR-29b-3p. We also used the ENCORI database to examine the relationship between CDCA4 and lncRNAs and miRNAs in LIHC.
Gene set enrichment analysis (GSEA)
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted to investigate the biological roles of CDCA4 in LIHC. The cellular component (CC), biological process (BP), and molecular function (MF) annotations associated with CDCA4 were determined using the powerful bioinformatics tool in the GO analysis. The mechanism of CDCA4 was investigated by a GSEA.
Statistical analysis
A GSEA was conducted to examine the probable cellular mechanism for CDCA4, and the statistical analysis and visualization were performed in R (version 3.6.3). (https://www.r-project.org/). The patients’ survival rates were calculated using the KM technique, and statistical significance was determined using the log-rank test. A P<0.05 indicated a statistically significant difference.
Results
Expression analysis of CDCA4
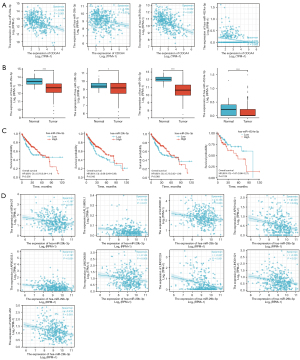
In this study, the GTEX and TCGA databases were used to investigate the CDCA4 mRNA expression levels in 33 different tumor tissues and adjacent tissues. We found that the CDCA4 mRNA expression levels varied across the tumor tissues and normal tissues, with certain tumor types [e.g., bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), colon adenocarcinoma (COAD), and kidney renal clear cell carcinoma (KIRC)] showing higher CDCA4 expression levels than others, and other tumor types [e.g., kidney chromophobe (KICH)] showing lower CDCA4 expression levels than others (Figure 1A). We also used TCGA database to analyze CDCA4 expression in cancer tissues paired with healthy tissues in patient malignancies. We discovered that BLCA, BRCA, cholangiocarcinoma (CHOL), and colon adenocarcinoma (COAD) tumor tissues had higher levels of CDCA4 expression than normal tissues, but KIRC tumor tissues had lower levels of the protein (Figure 1B). The association between the CDCA4 mRNA expression level and the OS of the pan-tumor patients in TCGA database was further assessed by the log-rank test and a KM survival analysis. According to the findings, cancers with CDCA4 mRNA lower expression levels of adrenocortical carcinoma (ACC), BLCA, head and neck squamous cell carcinoma (HNSC), kidney renal papillary cell carcinoma (KIRP), and acute myeloid leukemia (LAML) had a better prognosis (Figure 1C).
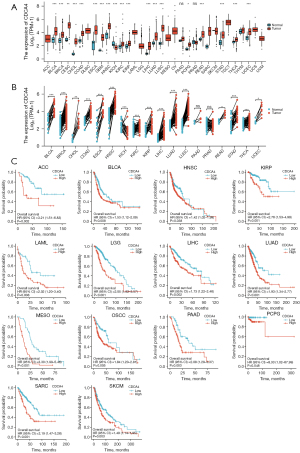
Upregulation of CDCA4 expression in LIHC tumor tissues
The expression of CDCA4 was found to be elevated in both the paired and unpaired LIHC tumor tissues in TCGA database (Figure 2).

CDCA4 overexpression is associated with adverse clinical parameters in LIHC patients
The results of our correlation analysis of CDCA4 expression and the clinical characteristics of LIHC patients showed that the LIHC patients with high CDCA4 expression had alpha-fetal protein (AFP) value is greater than 400 ng/mL, pathological grade, lymph node (N) stage, pathological stage, tumor (T) stage, vascular invasion, nutritional condition, etc. (see Figure 3).

The diagnostic and prognostic value of CDCA4
We used a KM survival analysis and the log-rank test to analyze the correlation between CDCA4 mRNA expression level and OS, disease-specific survival (DSS), and the progression free interval (PFI) of the LIHC patients in TCGA database. The LIHC patients with low expression levels of CDCA4 had improved OS, DSS, and PFI (Figure 4A). CDCA4 expression level was reported to have a reasonably high performance in predicting the 1-, 3-, and 5-year OS of LIHC patients based on the time-dependent receiver operating characteristic (ROC) curve (Figure 4B).
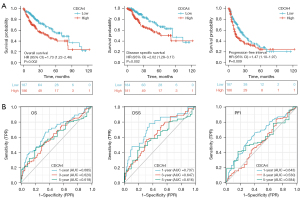
Predictive value of CDCA4 in LIHC clinical subgroups
A subgroup stratification study was performed in TCGA database to further investigate the predictive value of CDCA4 in LIHC. We can find that patients with low expression of CDCA4 have better prognosis in different LIHC subgroups (Figure 5).
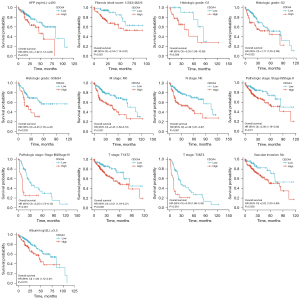
Pathway and gene function annotation analysis
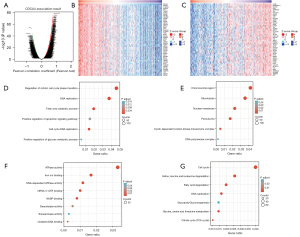
Subsequently, we used LinkedOmics to obtain genes significantly related to CDCA4 gene expression The heatmap results showed 50 gene sets that had substantial positive or negative relationships with CDCA4 (Figure 6A-6C). Next, we conducted GO and KEGG analyses to demonstrate that these genes regulate mitotic cell cycle phase transition, DNA replication, fatty acid catabolism, and the positive regulation of apoptosis signaling pathway, etc. (Figure 6D). Analysis of biological process and molecular function of CDCA4 shows that it is related to chromosome region, cyclin-dependent protein kinase holoenzyme complex, iron ion binding, nicotinamide adenine dinucleotide phosphate (NADP) binding, transaminase activity and oxidative DNA binding (Figure 6E,6F). The KEGG molecular pathways included the cell cycle, valine, leucine, and isoleucine degradation, fatty acid degradation, DNA replication, glycolysis/glucogenesis, glycine, serine, and threonine metabolism, and citrate cycle (Figure 6G).
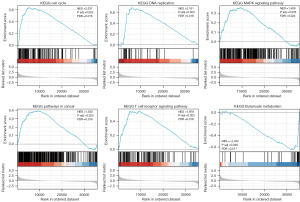
CDCA4 signaling pathways based on the GSEA analysis
We compared the median CDCA4 expression levels across the low and high-expression groups to examine the biological role of CDCA4. The GSEA showed that CDCA4 was primarily involved in the cell cycle, T cell receptor signaling pathway, DNA replication, glucose metabolism, and mitogen activated protein kinase (MAPK) signaling pathway (Figure 7).
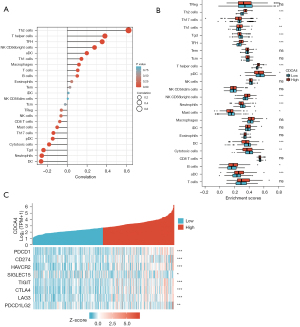
The correlation among CDCA4 expression and immunological infiltration and immune checkpoints
Lymphocytes that infiltrate tumors are crucial to cancer development and may change the prognosis of LIHC patients. Thus, we next examined whether CDCA4 was correlated with the level of immune infiltration in LIHC. We found a negative correlation between CDCA4 mRNA expression and dendritic cell (DC), neutrophils, Tgd, and plasma cell like dendritic cells (Pdc), but a positive correlation between CDCA4 and Th2 cells, T helper cells, follicular helper T cell (TFH), activated dendritic cells (aDC), and T helper cell 1 (Th1 cells), etc. (Figure 8A). The probable oncogenic role of CDCA4 in LIHC prompted us to examine its association with other genes involved in the disease. Given that CDCA4 may be a potential oncogene of LIHC, the correlation between CDCA4 and hepatitis a virus cellular receptor 2 (HAVCR2), lymphocyte activating 3 (LAG3), T cell immunoreceptor with Ig and ITIM domains (TIGIT), sialic acid binding Ig like lectin 15 (SIGLEC15), programmed cell death 1 (PDCD1), cytotoxic T-lymphocyte associated protein 4 (CTLA4), CD274 molecule (CD274), and programmed cell death 1 ligand 2 (PDCD1LG2) in LIHC was evaluated, and we found that CDCA4 was strongly correlated with these molecules (Figure 8B,8C), which suggests that CDCA4-mediated LIHC carcinogenesis may include immune evasion and anti-tumor immunity.
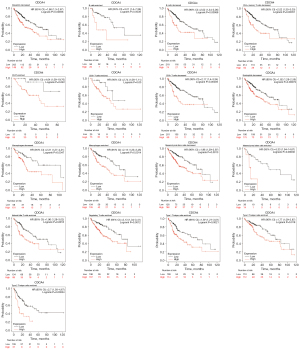
Prognostic value of CDCA4 in immune cell-based LIHC
According to the data presented above, LIHC patients with high expressions of CDCA4 have a poor prognosis. CDCA4 has been linked to the immune infiltration of LIHC. We found that among the rich CD4+ memory T cells, CD8+T cells, macrophages and eosinophils, the higher level of CDCA4 in LIHC had a poor prognosis (Figure 9).
Upstream CDCA4 miRNA and lncRNA analyses
There is mounting evidence that ncRNAs control gene expression at almost every stage. It is possible that CDCA4 is regulated by a set of ncRNAs. We predicted the upstream miRNAs that would bind to CDCA4 and ultimately identified 125 miRNAs. We predicted that CDCA4 and the upstream miRNAs would be inversely associated due to the mechanism through which the upstream miRNAs reduce CDCA4 production in the post-transcriptional stage. CDCA4 was shown to be correlated with 125 different miRNAs in the TCGA-LIHC database. Thus, we discovered that hsa-miR-29a-3p, hsa-miR-29b-3p, hsa-miR-29c-3p, and hsa-miR-4524a-5p were all significantly and adversely linked with the CDCA4 expression level (Figure 10A). Our subsequent analysis of the TCGA-LIHC database revealed that the expression of these 4 miRNAs was significantly lower in tumor tissues than normal tissues (Figure 10B). Next, using TCGA database, we investigated the link between these 4 miRNAs and the outcomes of LIHC patients. A high expression of hsa-miR-29b-3p was correlated with a better prognosis in LIHC patients, and there was no statistically significant difference in the correlation between the other three miRNAs and prognosis (P>0.05) (Figure 10C). Based on the combined findings of our correlation, expression, and survival analyses, we hypothesized that hsa-miR-29b-3p is the miRNA primarily responsible for regulating CDCA4 in LIHC.
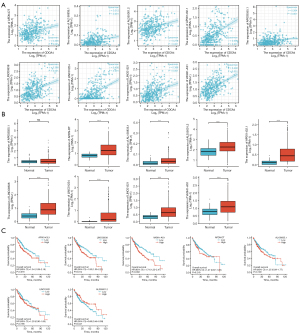
The predicted hsa-miR-29b-3p upstream lncRNA interactors were found using the ENCORI database. We narrowed the pool of potential lncRNAs down to 56 lncRNAs that interact with homo-sapiens microRNA (hsa-miR). The competing endogenous RNA (ceRNA) hypothesis states that lncRNAs compete with tumor suppressor microRNAs for binding, thereby decreasing the miRNA regulation of the target mRNAs. Thus, in the ceRNA network, lncRNAs should be negatively correlated with the target miRNAs and positively correlated with the target mRNAs. We then examined the role of these lncRNAs in LIHC in terms of both expression and prognosis. After careful consideration of the correlation, expression, and survival analyses, we found that lncRNA-AP001432.1, LINC00638, and VASH1-AS1 were the potential regulatory lncRNAs of hsa-miR-29b-3p (Figure 10D, Figure 11A-11C). Based on our analysis and the literature, we concluded that LINC00638 is the most plausible upstream lncRNA of the CDCA4/hsa-miR-29b-3p axis in LIHC.
Discussion
According to the global cancer statistics report, the morbidity and mortality of LIHC rank sixth and fourth, respectively (14,15). The screening of new LIHC diagnostic genes and prognostic markers is of great significance in improving the prognosis of LIHC patients. In this study, we used TCGA and GTEX data sets to examine the mRNA expression levels of CDCA4 in pan-tumor and comparable surrounding normal tissues. When we compared the results of our expression study to those of a validation analysis, we found that CDCA4 was significantly overexpressed in most tumor tissues. In the LIHC tissues, we observed an upregulation of CDCA4 mRNA, which was associated with worse clinical outcomes. The ROC analysis suggested that CDCA4 might be a useful diagnostic biomarker for identifying LIHC tissues as opposed to normal tissues. The KM curve analysis showed that TCGA patients with LIHC who expressed low levels of CDCA4 had better OS, DSS, and PFI.
According to previous research, CDCA4 is a protein-coding gene that belongs to the E2F transcription factor (E2F) family and may regulate the transcriptional activity of target genes, including tumor protein P53 (p53), E2F, and Jun proto-oncogene (JUN, AP-1 transcription factor subunit), and thus plays an important role in the cell cycle, proliferation, and apoptosis (16). In this study, we examined the underlying processes by which CDCA4 affects LIHC development. According to the GSEA results, CDCA4 is mostly involved in the cell cycle, T cell receptor signaling route, DNA replication, glucose metabolism, and MAPK signaling pathway, all of which have an effect on the BPs of LIHC. We also showed that the mRNA expression level of CDCA4 was significantly positively correlated with Th2 cells, T helper cells, TFH, aDC, and Th1 cells, etc., but was negatively correlated with DC, neutrophils, Tgd, and pDC. CDCA4 has a good correlation with HAVCR2, LAG3, TIGIT, CD274, PDCD1, CTLA4, SIGLEC15, and PDCD1LG2. These findings imply that CDCA4-mediated LIHC carcinogenesis may include tumor immune evasion and anti-tumor immunity.
With the progress of medical research, immunotherapy for LIHC is currently in full swing. The tumor immune microenvironment, that is, a large number of immune cells are often gathered inside and around the tumor. These immune cells have complex interactions and regulation with tumor cells. Therefore, when we are looking for biomarkers available for tumors, we hope that this molecule can not only predict the prognosis of patients, but also be related to the tumor’s immune microenvironment, so as to provide a reference therapeutic target for future tumor immunotherapy. In this study, we discovered that CDCA4 may partially affect the prognosis of LIHC patients via immune infiltration.
To further reveal the potential upregulation of CDCA4 in LIHC, we conducted correlation analysis, expression analysis, and survival analysis of these miRNAs in LIHC. We discovered an inverse relationship between CDCA4 and hsa-miR-29b-3p expression levels, and found that CDCA4 was more lowly expressed in tumor tissues than normal tissues. A positive prognosis after LIHC was found to be significantly correlated with high levels of hsa-miR-29b-3p expression in the survival study. Since upstream miRNAs suppress CDCA4 production at the post-transcriptional stage, the ceRNA hypothesis (17,18) predicted an inverse relationship between CDCA4 and the upstream miRNAs. Evidence from multiple aspects of research, including the ceRNA hypothesis, expression, correlation, and survival analyses, led us to conclude that hsa-miR-29b-3p is a strong candidate for the miRNA that regulates CDCA4 in LIHC. According to our results, hsa-miR-29b-3p may inhibit LIHC by targeting CDCA4.
By searching the ENCORI database, we were able to narrow down the pool of possible interacting lncRNAs to 56 lncRNAs upstream of hsa-miR-29b-3p. The ceRNA hypothesis states that the putative lncRNA may have a positive association with CDCA4, a negative correlation with hsa-miR-29b-3p, and it should be an oncogenic lncRNA in LIHC. We discovered the following 3 potential regulating lncRNAs for hsa-miR-29b-3p in the correlation, survival, and expression analyses: lncRNA-AP001432.1, LINC00638, and VASH1-AS1. Previous studies have reported that lncRNA-AP001432.1 is an independent prognostic indicator in patients with soft tissue sarcoma (19); LncRNA-VASH1-AS1 is currently only useful in comprehensive research on the effect of radiotherapy for rectal tumors (20); the overexpression of lncRNA LINC00638 inhibits inflammation and oxidative stress in rheumatoid arthritis fibroblast-like synoviocytes by regulating the NFE2 like BZIP transcription factor 2/heme oxygenase 1 (Nrf2/HO-1) pathway (21); the low expression of lncRNA Linc00638 promotes the progression of rheumatoid arthritis by modulating inflammation and oxidative stress (22); immune escape through PD-L1 (CD274 molecule) is facilitated in hepatocellular carcinoma by the LINC00638/miR-4732-3p/UL16 binding protein 1 (ULBP1) axis, which is correlated with the tumor mutational load (23). LINC00638 is the most likely upstream lncRNA on the CDCA4/hsa-miR-29b-3p axis in LIHC. In summary, we believe that LINC00638/hsa miR-29b-3p/CDCA4 should be a potential regulatory pathway in LIHC.
Conclusions
This study has improved our understanding of the correlation between CDCA4 and LIHC, but there are still some limitations. First, although we have explored the correlation between CDCA4 and immune infiltration in patients with LIHC, there is a lack of experiments to verify the role of CDCA4 in the regulation of tumor microenvironment in LIHC. At the same time, further multicenter clinical trials are needed to verify the clinical value of our research in the future.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-569/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-569/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-569/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Puccini A, Battaglin F, Iaia ML, et al. Overcoming resistance to anti-PD1 and anti-PD-L1 treatment in gastrointestinal malignancies. J Immunother Cancer 2020;8:e000404. [Crossref] [PubMed]
- Aizarani N, Saviano A. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019;572:199-204. [Crossref] [PubMed]
- Selçuk H. Prognostic Factors and Staging Systems in Hepatocellular Carcinoma. Exp Clin Transplant 2017;15:45-9. [PubMed]
- Grohmann M, Wiede F, Dodd GT, et al. Obesity Drives STAT-1-Dependent NASH and STAT-3-Dependent HCC. Cell 2018;175:1289-1306.e20. [Crossref] [PubMed]
- Petrick JL, Kelly SP, Altekruse SF, et al. Future of Hepatocellular Carcinoma Incidence in the United States Forecast Through 2030. J Clin Oncol 2016;34:1787-94. [Crossref] [PubMed]
- Abdullah JM, Jing X, Spassov DS, et al. Cloning and characterization of Hepp, a novel gene expressed preferentially in hematopoietic progenitors and mature blood cells. Blood Cells Mol Dis 2001;27:667-76. [Crossref] [PubMed]
- Walker MG. Drug target discovery by gene expression analysis: cell cycle genes. Curr Cancer Drug Targets 2001;1:73-83. [Crossref] [PubMed]
- Watanabe-Fukunaga R, Iida S, Shimizu Y, et al. SEI family of nuclear factors regulates p53-dependent transcriptional activation. Genes Cells 2005;10:851-60. [Crossref] [PubMed]
- Tao Q, Chen S, Liu J, et al. The roles of the cell division cycle-associated gene family in hepatocellular carcinoma. J Gastrointest Oncol 2021;12:781-94. [Crossref] [PubMed]
- Hu J, Xiang X, Guan W, et al. MiR-497-5p down-regulates CDCA4 to restrains lung squamous cell carcinoma progression. J Cardiothorac Surg 2021;16:330. [Crossref] [PubMed]
- Liu J, Tao G, Zhong C, et al. Upregulation of miR-29c-3p Hinders Melanoma Progression by Inhibiting CDCA4 Expression. Biomed Res Int 2021;2021:7065963. [Crossref] [PubMed]
- Li J, Zhang F, Li H, et al. Circ_0010220-mediated miR-503-5p/CDCA4 axis contributes to osteosarcoma progression tumorigenesis. Gene 2020;763:145068. [Crossref] [PubMed]
- Zhang J, Xia S, Liu X, et al. STAT1 Mediates the Transcription of CircIFI30 and Promotes the Progression of Triple-Negative Breast Cancer by Up-Regulating CDCA4. J Environ Pathol Toxicol Oncol 2022;41:1-13. [Crossref] [PubMed]
- Shah P, Wolf K, Lammerding J. Bursting the Bubble - Nuclear Envelope Rupture as a Path to Genomic Instability? Trends Cell Biol 2017;27:546-55. [Crossref] [PubMed]
- Ohnuma S, Miura K, Horii A, et al. Cancer-associated splicing variants of the CDCA1 and MSMB genes expressed in cancer cell lines and surgically resected gastric cancer tissues. Surgery 2009;145:57-68. [Crossref] [PubMed]
- Fang H, Sheng S, Chen B, et al. A Pan-Cancer Analysis of the Oncogenic Role of Cell Division Cycle-Associated Protein 4 (CDCA4) in Human Tumors. Front Immunol 2022;13:826337. [Crossref] [PubMed]
- Yan P, Huang Z, Mou T, et al. Comprehensive analyses of competing endogenous RNA networks reveal potential biomarkers for predicting hepatocellular carcinoma recurrence. BMC Cancer 2021;21:436. [Crossref] [PubMed]
- Zhang L, Tao H, Li J, et al. Comprehensive analysis of the competing endogenous circRNA-lncRNA-miRNA-mRNA network and identification of a novel potential biomarker for hepatocellular carcinoma. Aging (Albany NY) 2021;13:15990-6008. [Crossref] [PubMed]
- He RQ, Wei QJ, Tang RX, et al. Prediction of clinical outcome and survival in soft-tissue sarcoma using a ten-lncRNA signature. Oncotarget 2017;8:80336-47. [Crossref] [PubMed]
- Kutilin DS, Gusareva MA, Kosheleva NG, et al. Regulatory network of competitively interacting RNAs and effectiveness of rectal tumors radiotherapy. Klin Onkol 2022;35:297-306. [Crossref] [PubMed]
- Sun Y, Liu J, Wen J, et al. Overexpression of long noncoding RNA LINC00638 inhibits inflammation and oxidative stress in rheumatoid arthritis fibroblast-like synoviocytes by regulating the Nrf2/HO-1 pathway. Immun Inflamm Dis 2022;10:e663. [Crossref] [PubMed]
- Sun Y, Liu J, Xin L, et al. The low expression of long non-coding RNA Linc00638 contributes to rheumatoid arthritis progression by regulating inflammation and oxidative stress. Nan Fang Yi Ke Da Xue Xue Bao 2021;41:965-71. [PubMed]
- Qi F, Du X, Zhao Z, et al. Tumor Mutation Burden-Associated LINC00638/miR-4732-3p/ULBP1 Axis Promotes Immune Escape via PD-L1 in Hepatocellular Carcinoma. Front Oncol 2021;11:729340. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


