
The clinical significance of loss of FHIT and PTEN expression in 289 patients with non-small-cell lung cancer
Introduction
Lung cancer remains the leading causes of cancer mortality both in men and in women and is the most common cancer both in incidence and in mortality globally (1.35 million deaths annually). Non-small-cell lung cancer (NSCLC) accounts for approximately 80% of lung cancers diagnosed (1). The long-term survival rate of patients remains unsatisfactory with NSCLC due to unclear pathological mechanism. Thus it urgent for us to further understand the molecular mechanisms of NSCLC and finding new molecular targets for treatment.
FHIT and PTEN genes are tumor suppressor gene reported many years ago, but research about the loss of FHIT and PTEN expression is still interesting and useful for clinic. According to literature reported, the FHIT gene is a nucleotide metabolism associated with the Ap3A hydrolase, which may regulate cell cycle and induce cell apoptosis (2). Also, PTEN gene is the first discovery of a dual specificity phosphatase activity of tumor suppressor gene (3). Frequent inactivation of the PTEN tumor suppressor gene leads to negatively regulating phosphatidylinositol 3 phosphate levels (4) and alterations of this gene have been identified in a large fraction of cancers including lung cancer (5). Thus, we sought to explore the role of FHIT and PTEN gene in lung tumorigenesis through using immunohistochemistry staining to detect the expression level of FHIT and PTEN gene in normal lung tissue and benign lung lesions, compared with NSCLC.
Methods
Patients
Clinical data and tissue samples were obtained from surgical specimens from July, 2008 to October, 2010. All patients gave informed consent before collection of the specimens. Institutional review board was approval for this study. Forty cases normal lung tissue and 36 cases benign pulmonary lesion tissues included 48 males and 28 females ranging from 19 to 68 years (median, 53 years) of age. Lung benign lesions were from adjacent normal lung tissue including pulmonary tuberculosis (10 cases), bronchiectasis (8 cases), inflammatory pseudotumor (5 cases), bronchogenic cysts (6 cases), pulmonary bullae (3 cases), hamartoma (4 cases). The 289 NSCLC patients consisted of 164 males and 125 females ranging from 26 to 82 years (median, 59.63 years) of age, including 151 smokers and 138 non-smokers; N0 (96 cases), N1-2 (193 cases). Histological types included squamous cell carcinomas (120 cases), adenocarcinomas (139 cases), adenosquamous carcinomas (22 cases), large cell carcinomas (8 cases); according to the degree of differentiation, these NSCLC patients were classified into poorly differentiated carcinomas (93 cases), differentiated carcinomas (122 cases), high differentiated carcinomas (74 cases). Histological types were managed according to the World Health Organization Histologic Typing of Lung Tumors. The pathological stages of the patients were based on the seventh TNM Classification of Malignant Tumors. Pulmonary resection was performed mainly for early disease stages [stage I (96 cases), stage II (91 cases), stage III(98 cases), stage IV (4 cases)]—including solitary brain metastasis (3 cases), solitary adrenal metastasis (1 case), all of which underwent gamma knife treatment before radical operation of lung cancer.
Follow up
Two hundred and eighty nine cases of NSCLC patients were followed up by telephone, letters and other forms. All the patients had complete record of follow-up. The deadline of follow-up was November 2014. The survival time was calculated from the date of finishing surgery to 60 months of follow-up or the date of death (any cause). A total of 105 patients survived until the deadline of follow-up.
Primary antibodies
The rabbit polyclonal anti-FHIT antibody (ZYMED BioTechnology Co. Ltd, USA) was used at a 1:200 dilution; Mouse monoclonal anti-PTEN (Zhongshan Jinqiao BioTechnology Co. Ltd., Beijing, China) was used at a 1:200 dilution.
Immunohistochemistry
Tissue samples were fixed in 4% formalin and were processed for paraffin embedding. Then, histological sections were cut with 4 mm thickness. The tissue samples were deparaffinized through a series of xylene baths and were rehydrated through graded alcohols. Using Immunohistochemical method of streptavidin-peroxidase (S-P) determines the protein expression of FHIT and PTEN, and using the high temperature water bath repairs the antigen. The positive control was a section from breast carcinoma, while the negative control staining was done with phate buffer saline (PBS). The sections were incubated with a primary antibody overnight at 4 °C, and the primary antibody was diluted at the ratio of 1:200. FHIT and PTEN gene expression products appeared in the patina or nucleus. When gene expression was positive in cytoplasm, some nuclear membrane showed brown granules.
Immunohistochemical scoring
According to the percent of positive cells, relevant score was given for each as follows: <5% of the cells =1 point; 5–50% of the cells =2 points; >70% of the cells =3 points. Another score was given according to the intensity of staining as follows (6): negative staining =0 point; weak staining (light yellow) =1 point; moderate staining (yellowish brown) =2 points; strong staining (brown) =3 points. A final score was then calculated by multiplying two kinds of scores previously. If the final score was >3, the tumor was considered positive expression; otherwise, the tumor was considered negative expression. The degree of immunostaining was reviewed and calculated by two independent observers.
Statistical analysis
All statistical analyses were conducted using the SPSS 17.0 statistical software package. Categorical variables were evaluated by cross tabulation and the χ2 test. A Chi-square test (Phi correlation) was perform to examine the relationship between FHIT and PTEN expression. Using Kaplan-Meier method calculates the overall patients survival rate. The difference in survival curves was evaluated by using a Log-rank test. The correlation between clinical and biological characteristics and survival were analyzed by univariate and multivariate Cox proportional hazard models. All of the tests were two-sided. A P value of <0.05 in all cases was considered statistically significant.
Result
FHIT and PTEN protein expression
FHIT and PTEN gene expression were primarily in the cytoplasm or nucleus. We detected the negative rate of FHIT and PTEN expression in 2 of 76 (2.63%) and 3 of 76 (3.94%) in normal lung specimens respectively, compared to the negative rate of FHIT and PTEN expression in NSCLC specimens 157 of 289 (54.32%) and 173 of 289 (59.86%) respectively (P<0.001) (Table 1). The negative rate of FHIT expression includes 67.50% (81/120) squamous cell carcinoma, 46.04% (64/139) adenocarcinoma, 45.45% (10/22) adenosquamous carcinoma, 25.00% (2/8) large cell carcinoma (Figure 1). The negative rate of PTEN expression includes 56.67% (68/120) squamous cell carcinoma, 64.02% (89/139) adenocarcinoma, 50.00% (11/22) adenosquamous carcinoma, 62.50% (5/8) large cell carcinoma (Figure 2). Compared to normal lung specimens, the negative rate of FHIT and PTEN expression were statistically significant (P<0.001, respectively) (Table 2).
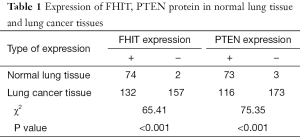
Full table
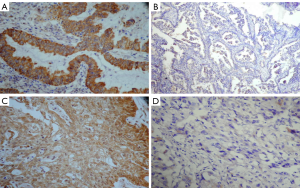
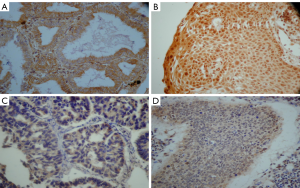
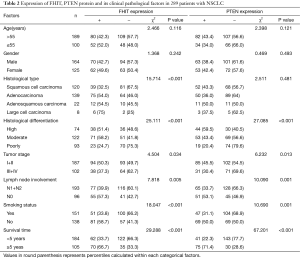
Full table
Correlation between the expressing levels of FHIT and PTEN protein and the clinical and pathological features of patients with NSCLC
Among the patients with NSCLC, the negative expression of FHIT was unrelated to age and gender. With regard to tumor characteristics, the loss of FHIT expression was more significantly found in histological type, poor differentiation, lymph node involvement, TNM-staging and smoking status. On the other hand, the negative expression of PTEN was unrelated to age, gender and histological type. With regard to tumor characteristics, the loss of PTEN expression was more significantly found in poor differentiation, TNM-staging, lymph node involvement, and smoking status (Table 2). A significant positive correlation (phi=0.199, P<0.001) was observed between FHIT and PTEN expression (Table 3). Therefore, we speculate that there has low correlation between these two genes lost together.
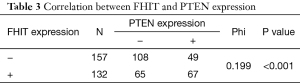
Full table
Expression of FHIT, PTEN and prognosis
At the end of follow-up, 105 NSCLC patients were still alive. Patients with negative expression of FHIT and PTEN showed significantly worse 5-years survival rate than those with FHIT and PTEN positive expression (33.33% vs. 66.67%, 28.57% vs. 71.43%, respectively, Figure 3). The result of multivariate Cox analysis showed that smoking, stage and FHIT and PTEN expression were independent prognosticators (Table 4).
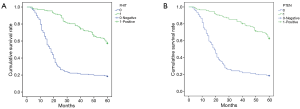
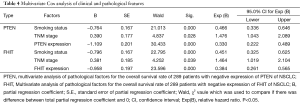
Full table
Discussion
The imbalance between oncogene and anti-oncogene is deemed to be the basis of tumorigenesis, FHIT and PTEN gene are tumor suppressor gene reported many years ago, which play an important role in the process of occurrence and development in lung cancer. The FHIT gene is located on 3P14.2, occupying with the 1Mb size of the position. This gene includes t (3;8) translocation and FRA3B brittle, and it is easily affected by external environment. The FHIT gene is the first molecular events associated with brittle point instability and tumor (2). PTEN gene is located on chromosome 10 in district 23.3, including 9 exons and 8 introns, full-length 200KB, exon 5 coding region with bispecific phosphatase function. The PTEN gene encodes a protein of 403 amino acids, possessing dual specificity phosphatase activity of protein. The PTEN protein is regulated by PI3PK/PKB signaling pathways and thereby functioned as inducing apoptosis and inhibiting cell growth (7,8). In addition, PTEN protein can directly work on the local adhesion kinase (FAK) and prevent tumor metastasis (9). Moreover, other studies showed that the inactivation of PTEN gene caused by mutation or deletion results in lower PTEN protein expression or no PTEN protein expression (4,5). The common PTEN mutation was in exon 5, 7 and 8, which is easily affected by the environment and other factors. What’s more, PTEN can cause the PI3P2 to remove 3—phosphate and inactivation, loss of messenger function, known as protein kinase B signal transduction pathway “switch” (10). Therefore, the deletion mutation of the PTEN gene or inactivation of the expression product might be closely associated with the genesis and development of cancer (11,12)
In our study, there were higher rates of loss of FHIT and PTEN expression in NSCLC specimens (54.32% and 59.86%, respectively) when compared to normal lung tissue (P<0.001), which indicate that this association may have implications for tumorigenesis. Moreover, several publications (13-15) have reported their result the same as our further analysis that loss of FHIT expression was related to squamous cell carcinoma and adenocarcinoma (P<0.001). According to previous literature reported, PTEN mutations were significantly more frequent in squamous cell carcinoma than in adenocarcinoma (16-18), while our study showed the loss of PTEN expression in adenocarcinoma and in squamous cell carcinoma without significance (64.03% vs. 56.67%, P>0.05). We consider that low statistical power may account for this observation. In addition, whether the mutation of PTEN gene will affect the expression of the protein needs further study.
Expression of FHIT and PTEN protein were related to differentiation of lung cancer, TNM-staging and prognosis. In our study, loss of FHIT and PTEN expression in early stage lung cancer (stage I + II) were 49.73% (93/187) and 54.55% (102/187), while 62.75% (64/102) and 69.61% (71/102) in advanced lung cancer (stage III + IV) (P=0.034, P=0.013, respectively). Moreover, patients with loss of FHIT and PTEN expression showed significantly worse 5 years survival rate than relevant those with FHIT and PTEN positive expression. Our findings support those of previously published reports that have suggested that smoking, TNM stage and FHIT and PTEN expression were independent prognosticators (19,20). Our result suggested that FHIT and PTEN might be an independent prognostic factor for patients with NSCLC. In addition, the detection of the expression of FHIT and PTEN may provide reference for postoperative radiotherapy and chemotherapy. Several studies have demonstrated that loss of PTEN expression contributes to gefitinib resistance in NSCLC (21,22).
According to our study, the loss of FHIT expression in smoking group was significantly higher than that of non smoking group (P<0.001). The FHIT gene contains the most common fragile site of human genome, FRE3B, which is easily induced to be broken by tobacco carcinogen (23,24). We can infer that tobacco, which contains many chemical mutagenesis, can induce gene deletion mutations and result in the loss of FHIT expression, which leads to the occurrence of tumor. FRA3B may be a preferential target of tobacco smoke damage at a molecular level and early molecular phenomenon of lung cancer. The expression of FHIT may be lower in squamous cell carcinoma due to its association with higher rates of smoking than adenocarcinoma. As for adenocarcinoma, the mechanism of loss expression of FHIT is still unclear. The connection between smoking and the loss expression of PTEN is also unclear and needs further study.
Conclusions
In summary, we have demonstrated that loss of FHIT and PTEN expression in clinical specimens could be invasion and metastasis of NSCLC. Our result suggested that loss of FHIT and PTEN expression confer poor prognosis in NSCLC. FHIT and PTEN are independent prognostic factor for patients with NSCLC. Restoring the imbalance of FHIT and PTEN expression may become a new target to treat NSCLC. In addition, tobacco can induce gene deletion mutations and result in the loss of FHIT expression.
Acknowledgments
We thank Mr Yang Li for his assistance with the data analysis.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.06.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional review board and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 1996;84:587-97. [Crossref] [PubMed]
- Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 1997;15:356-62. [Crossref] [PubMed]
- Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A 1999;96:4240-5. [Crossref] [PubMed]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell 2000;100:387-90. [Crossref] [PubMed]
- Pennisi E. New gene forges link between fragile site and many cancers. Science 1996;272:649. [Crossref] [PubMed]
- Planchon SM, Waite KA, Eng C. The nuclear affairs of PTEN. J Cell Sci 2008;121:249-53. [Crossref] [PubMed]
- Scrima M, De Marco C, Fabiani F, et al. Signaling networks associated with AKT activation in non-small cell lung cancer (NSCLC): new insights on the role of phosphatydil-inositol-3 kinase. PLoS One 2012;7:e30427 [Crossref] [PubMed]
- Zhang LL, Liu J, Lei S, et al. PTEN inhibits the invasion and metastasis of gastric cancer via downregulation of FAK expression. Cell Signal 2014;26:1011-20. [Crossref] [PubMed]
- Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal 2002;14:381-95. [Crossref] [PubMed]
- Hu TH, Huang CC, Lin PR, et al. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer 2003;97:1929-40. [Crossref] [PubMed]
- Mutter GL, Lin MC, Fitzgerald JT, et al. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J Natl Cancer Inst 2000;92:924-30. [Crossref] [PubMed]
- Chang X, Yang S, Zhang H, et al. Expression of FHIT gene in precancerous lesions and primary lung cancer tissues. Zhongguo Fei Ai Za Zhi 2006;9:320-4. [PubMed]
- Feng X, Li L, Gao Y, Zhang J, et al. Fhit protein expression in lung cancer studied by high-throughput tissue microarray. Bull Cancer 2007;94:E8-11. [PubMed]
- Woenckhaus M, Merk J, Stoehr R, et al. Prognostic value of FHIT, CTNNB1, and MUC1 expression in non-small cell lung cancer. Hum Pathol 2008;39:126-36. [Crossref] [PubMed]
- Li YN, Zhou YZ, Zhang N, et al. Expression of phosphatase and tensin homology deleted on chromosometen (PTEN) in squamous-cell lung cancer and its clinical significance. Zhonghua Jie He He Hu Xi Za Zhi 2016;39:450-3. [PubMed]
- Lee SY, Kim MJ, Jin G, et al. Somatic mutations in epidermal growth factor receptor signaling pathway genes in non-small cell lung cancers. J Thorac Oncol 2010;5:1734-40. [Crossref] [PubMed]
- Jin G, Kim MJ, Jeon HS, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer 2010;69:279-83. [Crossref] [PubMed]
- Tomizawa Y, Nakajima T, Kohno T, et al. Clinicopathological significance of Fhit protein expression in stage I non-small cell lung carcinoma. Cancer Res 1998;58:5478-83. [PubMed]
- Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol 2004;22:1878-85. [Crossref] [PubMed]
- Sos ML, Koker M, Weir BA, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res 2009;69:3256-61. [Crossref] [PubMed]
- Yamamoto C, Basaki Y, Kawahara A, et al. Loss of PTEN expression by blocking nuclear translocation of EGR1 in gefitinib-resistant lung cancer cells harboring epidermal growth factor receptor-activating mutations. Cancer Res 2010;70:8715-25. [Crossref] [PubMed]
- Tseng JE, Kemp BL, Khuri FR, et al. Loss of Fhit is frequent in stage I non-small cell lung cancer and in the lungs of chronic smokers. Cancer Res 1999;59:4798-803. [PubMed]
- D'Agostini F, Izzotti A, Balansky R, et al. Early loss of Fhit in the respiratory tract of rodents exposed to environmental cigarette smoke. Cancer Res 2006;66:3936-41. [Crossref] [PubMed]


