
Potential therapeutic targets for colorectal cancer and its subsites: evidence from the proteome-wide Mendelian randomization analyses
Highlight box
Key findings
• HHIP, FUT3, and CHRDL2 are potential therapeutic targets in colorectal cancer.
• IGF2R and CHRDL2 are potential therapeutic targets in colon cancer.
• ASRGL1 and CHRDL2 are potential therapeutic targets in rectal cancer.
What is known and what is new?
• Mendelian randomization method can effectively analyze the causal association between circulating proteins and tumors, and provide new therapeutic targets for tumor treatment.
• In this paper, we analyzed different anatomical sites and found that colon cancer and rectal cancer share common drug targets, but there are also differences.
What is the implication, and what should change now?
• Our study not only provides new drug targets, but also suggests the differences between colon cancer and rectal cancer. Different treatment options should be adjusted for colon cancer and rectal cancer in clinical treatment.
Introduction
Colorectal cancer (CRC) is the most common malignant tumor of digestive tract in the world, with the third incidence rate and the second mortality rate, which is a huge threat to human health (1). CRC has a hidden onset, and most patients are unable to undergo radical surgery when symptoms appear. The chemotherapy regimen for CRC is not satisfactory, and immunotherapy can only benefit a limited number of patients (2). Although endoscopic examination provides significant assistance for the early diagnosis of CRC, it is still undetectable even in the earliest stage (3). Therefore, developing new therapeutic targets for CRC is of great clinical significance.
Proteins are the direct targets of drugs. Plasma proteins exist in circulation, as drug targets, plasma proteins can act throughout body, providing hope for treatment for patients with advanced CRC metastasis. Plasma proteins come from various solid tissues, especially liver and intestines (4). This demonstrates the feasibility of developing drugs based on plasma proteins for CRC. Given the strong correlation between plasma proteins and tumors, targeting plasma proteins for tumor treatment is highly feasible. Currently, a large number of proteomic studies have confirmed the association between plasma proteins and CRC (5,6). However, these studies generally have small sample size and cannot rule out confounding factors and reverse causal effects. Genome-wide association studies (GWAS) have confirmed thousands of protein quantitative trait loci (pQTLs) (7). pQTLs are a series of single nucleotide polymorphism (SNP) sites strongly correlated with plasma protein levels. Based on pQTLs, we can evaluate the causal relationship between plasma proteins and diseases through Mendelian randomization (MR) methods, thereby identifying potential drug targets. Current study has shown that MR research has certain clinical guiding value (8,9).
Because colon originates from mesoderm and rectum originates from cloaca, the difference in embryology origin makes the two essentially different. There are also differences in treatment methods between colon cancer and rectum cancer. Develop specific drugs for different anatomical sites of colon cancer is an urgent problem to be solved. We selected a large population GWAS dataset for CRC as discovery cohort, FinnGen’s GWAS dataset for CRC as replication cohort, and further proteomic MR analysis was conducted on the plasma proteome dataset from Iceland. A proteomic analysis based on plasma of 83 CRC patients showed significant differences in plasma protein components between colon cancer and rectal cancer, and the protein phenotypes between left and right colon cancer are also different. Therefore, exploring treatment targets for CRC based on different anatomical sites is of great significance (10). Therefore, we extracted data from CRC, colon cancer, and rectal cancer separately and divided them into three groups. Finally, six proteins which have causal associations with CRC were identified. We present this article in accordance with the STROBE-MR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1503/rc).
Methods
MR design
The specific MR design is shown in Figure 1. This MR study uses STROBE-MR (STrengthening the Reporting of OBservational studies in Epidemiology-Mendelian Randomization) (https://www.strobe-mr.org/) guidelines for reporting, and it should follow three basic assumptions: (I) relevance criteria: genetic instruments should be closely related to exposure; (II) independence criteria: genetic variation should not be associated with any potential confounding factors that may mediate from exposure to outcome; (III) exclusion-restriction criterion: if being conditioned on exposure, genetic variation should not be associated with the outcome (11). This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).

Proteomic data source
There were 490,373 genetic variants associated with 4,907 plasma proteins (SomaScan version 4) obtained in a GWAS with 35,559 Icelanders (7). In further analysis, only pQTL that were genome-wide significant were kept (P value <5×10−8) and they were further clumped based a threshold of linkage disequilibrium (LD r2=0.01) (table available at https://cdn.amegroups.cn/static/public/TCR-24-1503-1.xlsx).
Outcome data sources
GWAS data of CRC in discovery cohort were obtained from the GWAS study with the largest sample size, which included 6,581 European patients, 8,305 East Asian patients, 463,421 European controls, and 159,386 East Asian controls (12). GWAS data for colon and rectal cancer in discovery cohort were obtained from a GWAS meta-analysis of two large cohorts [UK Biobank + Genetic Epidemiology Research on Adult Health and Aging (GERA)], including 3,793 European colon cancer patients, 2,091 European rectal cancer patients, and 410,350 European controls (13). GWAS of replication cohort comes from FinnGen (https://www.finngen.fi/en). All patients are of European ancestry, including 5,458 cases of CRC (3,292 cases of colon cancer and 2,017 cases of rectal cancer) and 259,583 controls (Table S1). Because of some patients’ tumor located in the borderline or multiple, the primary location cannot be determined and can only be classified as CRC.
MR analysis
We conducted a two-sample MR analysis based on pQTLs, using the Wald ratio method for proteins with only one SNP, and multiple random-effects inverse variance weighting (IVW) for proteins with more than one SNP. We calculated the odds ratio (OR) and corresponding confidence interval (CI) for the correlation between proteins and CRC and its subsites. In addition, weighted median was also used as a supplementary method. We conducted MR-Steiger directionality test to guarantee that the pQTL explained more variance of a protein than that of CRC. For the robustness of the results, a sensitivity test and heterogeneity test were also conducted by MR-Egger regression and MR-PRESSO. False discovery rate (FDR) was used to adjust multiple tests. Proteins with FDR <0.05 indicate a significant causal relationship with CRC and its subsites in the discovery stage and the protein was successfully replicated if its MR P value was <0.05 in the replication stage.
Colocalization analysis
We used the “coloc” R package to perform colocalization analysis to detect whether proteins share causal variations with CRC, colon cancer, and rectal cancer in separate regions of the genome (14). This analysis was established on five hypotheses. H0: no genetic association with either trait; H1: genetic association with only trait 1; H2: genetic association with only trait 2; H3: genetic association with both trait 1 and trait 2, but via different mechanism; H4: shared genetic association between two traits. The posterior probability (PP) of each hypothesis is calculated using the Bayesian method. Proteins with PPH4 >80% are considered to have a high colocalization association with outcomes, meaning that proteins directly affect outcomes.
Enrichment analysis and drug prediction
We further analyzed the associations between gene region and phenotypes through “ieugwasr” R package to find correlated phenotypes that might be associated with the SNPs in the genetic region that encodes the protein. We used WebGestalt (15) to perform enrichment analysis to predict protein regulatory pathways. In order to more effectively analyze the impact of proteins in CRC, we divided proteins obtained from MR results into the following two groups: OR >1 and OR <1, to identify pathways with more target proteins. Then, we searched the potential drugs pertaining to the identified proteins through DrugBank.
Statistical analyses and data visualization
All statistical analyses and data visualization were carried out using R package, including “TwoSampleMR”, “MRPRESSO”, “Coloc”, “miamiplot”, “ggplot2” and “locuscomparer” in R 4.2.1 (https://www.r-project.org/).
Results
MR analysis
We analyzed the association between protein pQTLs and CRC and its subsites, and conducted validation in replication cohort. Finally, proteins with FDR <0.05 in two cohorts were retained.
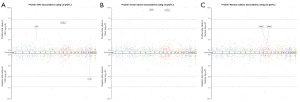
In CRC cohort, we found 31 related proteins based on 1,367 cis-pQTLs (Figure 2A, Table S2) and 29 related proteins based on 1,579 trans-pQTLs (Table S3). In colon cancer cohort, we found 23 related proteins based on 1,367 cis-pQTLs (Figure 2B, Tables S4) and 7 related proteins based on 1,579 trans-pQTLs (Table S5). In rectum cancer cohort, we found 23 related proteins based on 1,367 cis-pQTLs (Figure 2C, Tables S6) and 19 related proteins based on 1,579 trans-pQTLs (Table S7). Due to the low predictive power of trans-pQTLs, we only present the results of cis-pQTLs, while data of trans pQTLs is presented in the supplementary materials. In order to further screen more reliable protein targets, we further screened proteins with a higher degree of correlation with CRC and its subsites through colocalization analysis.
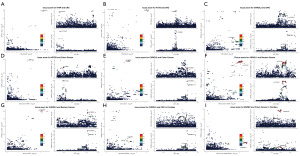
Colocalization analysis
We conducted colocalization analysis between selected plasma proteins and CRC and its subsites (table available at https://cdn.amegroups.cn/static/public/TCR-24-1503-2.xlsx), retaining proteins only with PPH4 >80%. We found three highly co-located proteins hedgehog interacting protein (HHIP), fucosyltransferase 3 (FUT3), and chordin-like 2 (CHRDL2) in CRC (Figure 3A-3C), two highly co-located proteins insulin-like growth factor 2 receptor (IGF2R) and CHRDL2 in colon cancer (Figure 3D,3E), and two highly co-located proteins asparaginase-like protein 1 (ASRGL1) and CHDRL2 in rectal cancer (Figure 3F,3G). Among them, CHRDL2 showed high colocalization in all three groups. In FinnGen, CHRDL2 also showed high colocalization with CRC (Figure 3H). In addition, we also found high colocalization between ENPEP and colon cancer in FinnGen (Figure 3I). These proteins are potential targets of CRC and subgroups with high level reliability. We also conducted the next functional analysis of these proteins.
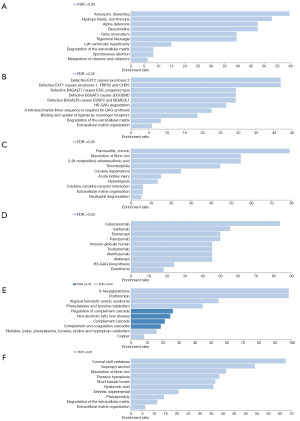
Enrichment analysis
We analyzed the functions of these proteins through WebGestalt, and get enrichment parameters of target genes in KEGG_pathway, Reactome, disease_Disgenet, disease_OMIM, drug_DrugBank five databases. In order to more effectively analyze the impact of circulating proteins on CRC and its subsites, we divided the proteins obtained from MR results into the following two groups: OR >1 and OR <1 Enrichment analysis was conducted separately to identify more pathways with target proteins (Figure 4A-4F) (associated data are in Tables S2,S4,S6). In CRC, selected genes were involved in the degradation and organization of extracellular matrix. In colon cancer, selected genes were involved in immune regulating. In rectum cancer, selected genes were involved in amino acids metabolism.
We analyzed the functions of these protein related SNPs through ieugwasr (Tables S8-S11 and tables available at https://cdn.amegroups.cn/static/public/TCR-24-1503-3.xlsx). PheWAS analysis shows these six proteins are involved in the formation of a large number of phenotypes, and it is worth mentioning that these genes are all involved in the regulation of lipid metabolism, which also indicates the key role of lipid metabolism in the treatment of CRC.
We screened drugs related to these proteins through DrugBank. We found 6 drugs targeted IGF2R and 4 of them have been approved. ENPEP have 1 targeted drug and ASRGL1 have 2 targeted drugs, these 3 drugs have been approved as nutraceutical (Table S12).
Discussion
Plasma molecules have important clinical potential for early diagnosis and treatment of colon cancer (16,17). We conducted a proteome-wide two-simple MR and explored the causal effect of 4,907 proteins in CRC, and found 31 proteins related to CRC, 23 proteins related to colon cancer, and 23 proteins related to rectal cancer. Colocalization analysis finally found 6 proteins highly associated with CRC. CHDRL2 was found to be positively correlated with the risk of CRC, colon cancer, and rectal cancer. HHIP was found to be positively correlated with the risk of CRC. FUT3 was found to be negatively correlated with the risk of CRC, IGF2R was found to be positively correlated with the risk of colon cancer, ENPEP was found to be positively correlated with the risk of colon cancer, and ASRGL1 was found to be positively correlated with the risk of rectal cancer.
CHRDL2
CHRDL2 is a protein of the Chordin family which is a secreted protein. CHRDL2 is an antagonist of bone morphogenetic protein 9 (BMP-9) and can prevent interactions between BMP-9 and its homologous cell surface receptors (18). Current research has confirmed that CHRDL2 is highly expressed in various tumor cells and has a cancer promoting effect. In CRC, high levels of CHRDL2 are closely related to poor prognosis. In vitro experiments, CHRDL2 binds to BMPs and inhibits p-Smad1/5, thereby promoting CRC cell proliferation and inhibit apoptosis (19). In osteosarcoma, CHRDL2 can activate PI3K/Akt pathway by downregulating BMP-9 and reducing binding between BMP-9 and ALK-1, thereby promoting proliferation and metastasis of osteosarcoma (20). In gastric cancer, CHRDL2 is highly expressed in tumor tissue and plasma, and is closely related to poor prognosis of gastric cancer. In vitro experiments, CHRDL2 promotes the proliferation of gastric cancer through the MST2/YAP/TAZ pathway (21). Our result strongly suggests that CHRDL2 can increase risk of CRC, colon cancer and rectum cancer in both discovery cohort and replication cohort. The cancer promoting function of CHRDL2 in tumors has been confirmed, but currently clinical research and drug development for this protein are still in a blank stage. CHRDL2 is a CRC target with excellent diagnostic and therapeutic potential.
HHIP
HHIP is a protein of Hedgehog (Hh) family which is evolutionarily conserved. The Hh pathway is a classic tumor promoting pathway, and HHIP has the function of inhibiting Hh pathway. Therefore, current research generally believes that HHIP is a tumor suppressor. CircFAM114A acting as competing endogenous RNA of miR-630, can upregulate HHIP and inhibit the progression of hepatocellular carcinoma (22). MiR-199b-5p can promote proliferation and metastasis of gastric cancer by inhibiting HHIP (23). However, we found a positive correlation between plasma levels of HHIP and risk of CRC. This may be related to the biological characteristics of CRC. Researchers have confirmed in 8 CRC cell lines that there is no abnormal activation of the Hh pathway or independent ligand mechanism activation. This may be why HHIP cannot play an inhibitory role in CRC. Other mechanisms of action of HHIP protein need further study (24). Currently there is no progress in drugs targeting HHIP, and researchers can consider HHIP as a specific drug target for CRC.
FUT3
FUT3 is associated with human tissue blood group antigens and secreted by the intestinal mucosa. FUT3 is a determining factor for Lewis blood type. A The Cancer Genome Atlas (TCGA) combined with Gene Expression Omnibus (GEO) analysis suggests that low expression of FUT3 is closely associated with poor prognosis in CRC (25). A large number of colon cancer cell lines study has shown that FUT3 can promote the differentiation level of colon cancer cells (26). High expression of FUT3 is commonly present in colon cancer cells sensitive to TRAIL, and FUT3 can activate DISC through indirect effects (27). However, upregulation of FUT3 accelerates TGF-β Fucose and active TGF-β signaling pathway ultimately drives the epithelial mesenchymal transition (EMT) and contributes to CRC progression (28). FUT3 is preferentially expressed in non-stem cell components of breast cancer and has the function of antagonizing tumor stem cells (29), deletion of FUT3 is also closely related to high invasiveness of breast cancer (30). FUT3 can promote tumor metastasis ability in non-small cell lung cancer (31). Our result suggests that FUT3 can reduce risk of CRC, while current research suggests that FUT3 may have a dual effect, but its mechanism of inhibiting CRC is clear and feasible. Therefore, protein therapy and drug development based on FUT3 in CRC may have a considerable potential.
IGF2R
IGF2R is a receptor for insulin-like growth factor 2 and mannose 6-phosphate, involved in intracellular transport of lysosomal enzymes and the transformation and activation of growth factors β and degradation of insulin like growth factor 2. The current omics research has confirmed that high expression of IGF2R is closely related to poor prognosis of patients with breast cancer (32), laryngeal cancer (33) and hepatocellular carcinoma (34). IGF2R is generally highly expressed in the plasma of tumor patients, which has considerable diagnostic efficacy (35). In non-small cell lung cancer, Trop2 binding to IGF2R promotes IGF2-IGF1R-Akt axis enhancement of gefitinib resistance (36). However, in cervical cancer, IGF2R can maintain lysosomal function, ensure autophagy and mitochondrial homeostasis of tumor cells, and maintain tumor cell survival (37). IGF2R plays an important regulatory role in autophagy and is an important factor in maintaining lysosomal function. The complex role of IGF2R in tumors is also related to the lysosomal autophagy pathway. IGF2R is also an effective target for treating tumors through the autophagy. Chemotherapy can increase expression of IGF2R on tumor cells surface, so developing chimeric antigen receptor T cell combined chemotherapy based on IGF2R has great therapeutic potential (38). At present, six drugs related to IGF2R have been developed, and we look forward to the future role of this target in combination therapy of colon cancer.
ENPEP
ENPEP encodes glutamyl aminopeptidase, which can cut off the N-terminal aspartate in angiotensin II, and is related to tumorigenesis and immune microenvironment. In a mixed study, it was found that patients with lower ENPEP expression had a higher response to immune checkpoint inhibitors (ICIs) treatment rates (39). TCGA data indicates that high expression of ENPEP is associated with poor prognosis in CRC (40). Our research suggests that ENPEP can increase risk of colon cancer. Currently, there is limited research on ENPEP in tumors, but a related drug has also been developed, and more research on this protein is needed in the future.
ASRGL1
ASRGL1 is an enzyme, which is initially an inactive precursor protein and becomes active after undergoing autocatalysis intramolecular treatment. The active form of ASRGL1 can catalyze the hydrolysis of L-asparagine and aspartate peptides (41). AGRSL1 can promote proliferation, invasion, and metastasis of hepatocellular carcinoma by inhibiting P53-CDK1 (42), high expression of AGRSL1 is also closely related to poor prognosis of hepatocellular carcinoma (40). In CRC, ASRGL1 is related to the immunosuppressive microenvironment and can be used as a prognostic marker (43). At present, there is little research on ASRGL1 in tumors. We propose that ASRGL1 is associated with an increased risk of rectal cancer. Currently, there are two drugs related to ASRGL1, and further research is needed on this target in the future.
One advantage of our study is that we used MR and colocation analysis to estimate the causal effect between circulating protein and CRC by using genetic variation. At the same time, we added group of colon cancer and rectal cancer to explore the differences between colon and rectum, two organs with different embryology origins. MR design minimizes deviations caused by confounding and reverse causal relationships, thereby improving causal reasoning. Colocalization analysis has been proven to be a powerful tool for revealing the pleiotropic effects of certain loci on multiple traits. In addition, we used GWAS with a large sample size, which increased the efficacy of detecting mild to moderate associations. In addition, we also validated it in the FinnGen. Another advantage is that we used the GWAS of the plasma proteome of the Icelandic population as instrumental variable, and the GWAS of CRC which is mostly European and some East Asian races, as the outcome, which makes our results more universal. There are some limitations in this study. First, horizontal pleiotropy cannot be minimized. Second, although we introduced data from Asian populations, the main body of this research is still European population which leads to a situation of population structure bias and limits the generalization to other population.
Conclusions
In summary, our study has identified CHRDL2, HHIP, and FUT3 as drug targets for CRC, CHRDL2, IGF2R, and ENPEP as drug targets for colon cancer, CHRDL2, and ASRGL1 as drug targets for rectal cancer. CHRDL2 is not only meaningful in CRC, but can also be detected in both discovery cohort and replication cohort, making it a highly promising drug target for CRC.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the STROBE-MR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1503/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1503/prf
Funding: This study was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1503/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Grothey A, Fakih M, Tabernero J. Management of BRAF-mutant metastatic colorectal cancer: a review of treatment options and evidence-based guidelines. Ann Oncol 2021;32:959-67. [Crossref] [PubMed]
- Patel SG, Karlitz JJ, Yen T, et al. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol 2022;7:262-74. [Crossref] [PubMed]
- Huang Z, Ma L, Huang C, et al. Proteomic profiling of human plasma for cancer biomarker discovery. Proteomics 2017; [Crossref] [PubMed]
- Surinova S, Choi M, Tao S, et al. Prediction of colorectal cancer diagnosis based on circulating plasma proteins. EMBO Mol Med 2015;7:1166-78. [Crossref] [PubMed]
- Bhardwaj M, Weigl K, Tikk K, et al. Multiplex quantitation of 270 plasma protein markers to identify a signature for early detection of colorectal cancer. Eur J Cancer 2020;127:30-40. [Crossref] [PubMed]
- Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 2021;53:1712-21. [Crossref] [PubMed]
- Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [Crossref] [PubMed]
- Lanlan C, Stephen B, Shan L, et al. First release of Mendelian randomisation book in Chinese. eGastroenterology 2023;1:e100043. [Crossref]
- Holm M, Joenväärä S, Saraswat M, et al. Plasma protein expression differs between colorectal cancer patients depending on primary tumor location. Cancer Med 2020;9:5221-34. [Crossref] [PubMed]
- Sanderson E, Glymour MM, Holmes MV, et al. Mendelian randomization. Nat Rev Methods Primers 2022;2:6. [Crossref] [PubMed]
- Sakaue S, Kanai M, Tanigawa Y, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet 2021;53:1415-24. [Crossref] [PubMed]
- Rashkin SR, Graff RE, Kachuri L, et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat Commun 2020;11:4423. [Crossref] [PubMed]
- Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014;10:e1004383. [Crossref] [PubMed]
- Liao Y, Wang J, Jaehnig EJ, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019;47:W199-205. [Crossref] [PubMed]
- Zhang H, Zhou J, Ye Y. Prediction and validation of circulating G-quadruplexes as a novel biomarker in colorectal cancer. J Gastrointest Oncol 2024;15:286-98. [Crossref] [PubMed]
- Chen L, Ma X, Dong H, et al. Construction and assessment of a joint prediction model and nomogram for colorectal cancer. J Gastrointest Oncol 2022;13:2406-14. [Crossref] [PubMed]
- Fujisawa T, Huang Y, Sebald W, et al. The binding of von Willebrand factor type C domains of Chordin family proteins to BMP-2 and Tsg is mediated by their SD1 subdomain. Biochem Biophys Res Commun 2009;385:215-9. [Crossref] [PubMed]
- Sun J, Liu X, Gao H, et al. Overexpression of colorectal cancer oncogene CHRDL2 predicts a poor prognosis. Oncotarget 2017;8:11489-506. [Crossref] [PubMed]
- Chen H, Pan R, Li H, et al. CHRDL2 promotes osteosarcoma cell proliferation and metastasis through the BMP-9/PI3K/AKT pathway. Cell Biol Int 2021;45:623-32. [Crossref] [PubMed]
- Wang L, Xu W, Mei Y, et al. CHRDL2 promotes cell proliferation by activating the YAP/TAZ signaling pathway in gastric cancer. Free Radic Biol Med 2022;193:158-70. [Crossref] [PubMed]
- Lai M, Li D, Liu M, et al. CircFAM114A2 inhibits the progression of hepatocellular carcinoma via miR-630/HHIP axis. Cancer Med 2023;12:12553-68. [Crossref] [PubMed]
- Chen S, Wu H, Zhu L, et al. MiR-199b-5p Promotes Gastric Cancer Progression by Regulating HHIP Expression. Front Oncol 2021;11:728393. [Crossref] [PubMed]
- Chatel G, Ganeff C, Boussif N, et al. Hedgehog signaling pathway is inactive in colorectal cancer cell lines. Int J Cancer 2007;121:2622-7. [Crossref] [PubMed]
- Wang P, Liu X, Yu J, et al. Fucosyltransferases Regulated by Fusobacterium Nucleatum and Act as Novel Biomarkers in Colon Adenocarcinoma. J Inflamm Res 2023;16:747-68. [Crossref] [PubMed]
- Wang D, Madunić K, Zhang T, et al. High Diversity of Glycosphingolipid Glycans of Colorectal Cancer Cell Lines Reflects the Cellular Differentiation Phenotype. Mol Cell Proteomics 2022;21:100239. [Crossref] [PubMed]
- Zhang B, van Roosmalen IAM, Reis CR, et al. Death receptor 5 is activated by fucosylation in colon cancer cells. FEBS J 2019;286:555-71. [Crossref] [PubMed]
- He C, Li A, Lai Q, et al. The DDX39B/FUT3/TGFβR-I axis promotes tumor metastasis and EMT in colorectal cancer. Cell Death Dis 2021;12:74. [Crossref] [PubMed]
- Walker MR, Goel HL, Mukhopadhyay D, et al. O-linked α2,3 sialylation defines stem cell populations in breast cancer. Sci Adv 2022;8:eabj9513. [Crossref] [PubMed]
- do Nascimento JC, Ferreira Sde A, Vasconcelos JL, et al. Fut3 role in breast invasive ductal carcinoma: Investigating its gene promoter and protein expression. Exp Mol Pathol 2015;99:409-15. [Crossref] [PubMed]
- Park S, Lim JM, Chun JN, et al. Altered expression of fucosylation pathway genes is associated with poor prognosis and tumor metastasis in non‑small cell lung cancer. Int J Oncol 2020;56:559-67. [PubMed]
- Zhong Y, Ren X, Cao X, et al. Insulin-like growth factor 2 receptor is a key immune-related gene that is correlated with a poor prognosis in patients with triple-negative breast cancer: A bioinformatics analysis. Front Oncol 2022;12:871786. [Crossref] [PubMed]
- Liu B, Hu Y, Wan L, et al. Proteomics analysis of cancer tissues identifies IGF2R as a potential therapeutic target in laryngeal carcinoma. Front Endocrinol (Lausanne) 2022;13:1031210. [Crossref] [PubMed]
- Lautem A, Simon F, Hoppe-Lotichius M, et al. Expression and prognostic significance of insulin‑like growth factor-2 receptor in human hepatocellular carcinoma and the influence of transarterial chemoembolization. Oncol Rep 2019;41:2299-310. [Crossref] [PubMed]
- Vicikova K, Petrovcikova E, Manka P, et al. Serum and urinary levels of CD222 in cancer: origin and diagnostic value. Neoplasma 2018;65:762-8. [Crossref] [PubMed]
- Sun X, Jia L, Wang T, et al. Trop2 binding IGF2R induces gefitinib resistance in NSCLC by remodeling the tumor microenvironment. J Cancer 2021;12:5310-9. [Crossref] [PubMed]
- Takeda T, Komatsu M, Chiwaki F, et al. Upregulation of IGF2R evades lysosomal dysfunction-induced apoptosis of cervical cancer cells via transport of cathepsins. Cell Death Dis 2019;10:876. [Crossref] [PubMed]
- Ramakrishnan R, Gabrilovich DI. The role of mannose-6-phosphate receptor and autophagy in influencing the outcome of combination therapy. Autophagy 2013;9:615-6. [Crossref] [PubMed]
- Wang A, Chu H, Jin Z, et al. ENPEP as a potential predictor of immune checkpoint inhibitor efficacy. Cancer Med 2022;11:880-7. [Crossref] [PubMed]
- Yuan Y, Chen J, Wang J, et al. Development and Clinical Validation of a Novel 4-Gene Prognostic Signature Predicting Survival in Colorectal Cancer. Front Oncol 2020;10:595. [Crossref] [PubMed]
- Biswas P, Chavali VR, Agnello G, et al. A missense mutation in ASRGL1 is involved in causing autosomal recessive retinal degeneration. Hum Mol Genet 2016;25:2483-97. [PubMed]
- Wang X, Wang Y, Yang L, et al. ASRGL1 downregulation suppresses hepatocellular carcinoma tumorigenesis in a CDK1-dependent manner. Dig Liver Dis 2023;55:955-66. [Crossref] [PubMed]
- Li F, Zhou J, Li Z, et al. Screening of immunosuppressive cells from colorectal adenocarcinoma and identification of prognostic markers. Biosci Rep 2021;41:BSR20203496. [Crossref] [PubMed]


