
METTL3 stabilizes SERPINE2 via the m6A modification to drive the malignant progression of gastric signet ring cell carcinoma
Highlight box
Key findings
• The study explored the role of serpin family E member 2 (SERPINE2) in the pathogenesis of gastric signet ring cell carcinoma (GSRCC).
What is known and what is new?
• SERPINE2 is highly expressed in gastric carcinoma, and its upregulation has relevance to aggressive phenotypes and forebodes poorer survival. However, SERPINE2 has not been defined in the context of GSRCC progression.
• Our study defines the significant roles of the novel methyltransferase-like 3 (METTL3)/SERPINE2 axis as an epigenetic mechanism in GSRCC progression.
What is the implication, and what should change now?
• The METTL3/SERPINE2 axis may be a potential target for the treatment of GSRCC.
Introduction
Despite of the low incidence rate (~10% of gastric carcinoma), gastric signet ring cell carcinoma (GSRCC) is a highly lethal, aggressive adenocarcinoma with poor prognosis (1). Clinically, treatment of GSRCC remains a huge challenge due to its strong invasiveness. Uncovering molecular determinants responsible for GSRCC development may provide encouraging therapeutic approaches for combating GSRCC (2). Abnormal angiogenesis can contribute to cancer progression, and anti-angiogenic agents have shown significant therapeutic potential in malignancy (3). Additionally, M2-polarized macrophages are potent inducers to pro-tumorigenic events, and inhibitors of M2 polarization have gained great attention due to their anti-cancer activity (4). Therefore, inhibitors of angiogenesis and M2 macrophage polarization may be efficient agents against GSRCC.
Serpin family E member 2 (SERPINE2) possesses anti-serine protease properties and thus functions as a suppressor of plasminogen, thrombin and urokinase (5). SERPINE2 is frequently overexpressed in many types of human malignancies, such as liver cancer and osteosarcoma (6,7). Overabundance of SERPINE2 is strikingly linked to worse outcomes of cervical and oral squamous cell carcinoma (8,9). Growing evidence highlights the pro-tumorigenic role of SERPINE2 in a broad spectrum of aggressive malignancies, including advanced renal cell carcinoma and liver cancer (6,10). In gastric carcinoma, SERPINE2 is highly expressed, and its upregulation has relevance to aggressive phenotypes and forebodes poorer survival (11). Recent work unveils the promoting impact of SERPINE2 on immunotherapeutic resistance in gastric carcinoma (12). Furthermore, SERPINE2 has not been defined in the context of GSRCC progression.
The expansion of knowledge of epigenetic dysregulation has uncovered significant roles in gastric tumorigenic processes in recent years (13). N6-methyladenosine (m6A) methylation, the most prevalent epigenetic RNA modification, is capable of controlling RNA metabolic processes, including RNA stabilization (14). Numerous studies have documented the critical implication of modification of RNA m6A methylation in gastric carcinogenesis (15,16). Moreover, the methyltransferase-like 3 (METTL3) has emerged as a critical player in gastric carcinoma development by mediating messenger RNA (mRNA) m6A methylation (17,18).
Here, we characterize SERPINE2 expression and function in GSRCC and point out the contribution of METTL3 to SERPINE2 upregulation and GSRCC progression. Our findings therefore suggest that the METTL3/SERPINE2 axis may help develop novel treatment candidates for GSRCC treatment. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-896/rc).
Methods
Computational analyses of GSRCC-related gene expression and survival association
To find out the differentially expressed genes (DEGs) in GSRCC, we utilized GSE33335 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33335) and defined DEGs with the criteria of P<0.05 and |log2foldchange| ≥1. We used Kaplan-Meier (KM)-plotter algorithm (http://kmplot.com/analysis/index.php?p=service) based on diffuse gastric cancer samples (n=241) to examine the association of DEGs and METTL3 with prognosis of GSRCC because GSRCC is a diffuse type of gastric carcinoma (1).
Collection of GSRCC samples and normal gastric tissues
We collected GSRCC tumors from a cohort of patients with GSRCC (n=35) treated at the Nuclear Industry 215 Hospital, Hospital of Shaanxi Province, as well as their paracancerous normal gastric tissues. Patients with other diseases were excluded. Fresh samples were rapidly frozen in liquid nitrogen or fixed in 4% formaldehyde. Before sample collection, patients were informed that their tissue specimens could be used for purpose of research. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Nuclear Industry 215 Hospital, Hospital of Shaanxi Province Ethics Committee (IRB No. 2023S29-UR1) and informed consent was taken from all the patients.
Cell lines
Human NUGC-4 (#C6717, Beyotime, Shanghai, China) and SNU-668 (#YS2933C, Yaji Biological, Shanghai, China) GSRCC cells and normal GES-1 cells (#IM-H084, Immocell, Xiamen, China) were cultured in RPMI-1640 (Invitrogen, Bleiswijk, the Netherlands) enriched with 10% fetal bovine serum (FBS, Invitrogen) and 1% antibiotic (100 U/mL penicillin and 100 µg/mL streptomycin) (Immocell) at 37 °C in an incubator containing 5% CO2. We obtained STR-authenticated human umbilical vein endothelial cells (HUVECs, #IM-H205) and human THP-1 monocytes (#IM-H260) from Immocell and cultivated them using standard complete mediums provided by Immocell. To induce differentiation to macrophages (THP-1/M0), THP-1 monocytes were maintained in complete media containing 100 ng/mL phytohaemagglutinin (PHA, Selleck, Shanghai, China) for 24 h. The medium was replaced with fresh medium every 2 days, and the cells were passed every 4–5 days.
Construction, transfection and transduction
To silence SERPINE2 in GSRCC cells, we obtained two shRNA constructs (sh-SERPINE2#1 and sh-SERPINE2#2) from VectorBuilder (Guangzhou, China), with a scrambled sh-NC control. The lentiviral vector pLV3-U6-METTL3(human)-shRNA-mCherry-Puro (sh-METTL3) was from Miaoling Biology (Wuhan, China) for in vitro knockdown of METTL3. The vector-based lentivirus generation was done by Genomeditech (Shanghai, China) for in vivo silencing of METTL3. We also purchased the pCMV-SERPINE2(human)-3×Flag-Neo construct from Miaoling Biology to express SERPINE2.
For plasmid transfection, we seeded NUGC-4 and SNU-668 cells 18 h before transfection to yield a density of 50–70% confluence at the transfection time. We generated liposomal cocktails with plasmid constructs and Lipofectamine 3000 (Invitrogen) in Opti-MEM (Invitrogen) as recommended by the supplier. Cells were maintained in media containing transfection complexes for 8 h before fresh media change, and transfected cells were cultured for 48 h prior to use. At the endpoint, we also harvested the conditioned medium (CM) from transfected NUGC-4 and SNU-668 cells for the subsequent use.
For virus transduction, we seeded SNU-668 GSRCC cells (106 cells per 10 cm dish) and incubated them overnight. At the time of infection, culture medium was replaced with 1:2, 1:3, or 1:4 diluted viral supernatant. After that, the incubation for 18–24 h was allowed. After 48 h infection, SNU-668 cells were subjected to puromycin selection (1 µg/mL, Selleck).
Generation of mouse xenografts
Experiments were performed under a project license (No. 6NI02-2023) granted by the Nuclear Industry 215 Hospital, Hospital of Shaanxi Province Animal Care and Use Committee, in compliance with national standard of the care and use of laboratory animals. We conducted in vivo xenograft experiments using BALB/c nude mice (female, n=10) (Vital River Laboratory Animal Technology Co., Ltd., Beijing, China). All animals were 6 to 8 weeks of age at the time of injection of SNU-668 GSRCC cells transduced with sh-NC (n=5) or sh-METTL3 (n=5) lentivirus (2×106 cells per mouse). Mice were randomly grouped using random number. The xenografts were assessed every five days for volume measurement, using the formula width2 × length × 1/2. At the time of necropsy, we collected the xenografts and recorded their weight. During experimental processes, dead mice were excluded.
Immunohistochemistry (IHC)
IHC analyses for SERPINE2, METTL3, or Ki67 probing in xenograft tumors were done as described earlier (19). In brief, sections (4 µm) of formalin-fixed paraffin-embedded tissues were deparaffinized, hydrated, and incubated with specific antibodies (Proteintech, Wuhan, China) recognizing SERPINE2 (rabbit polyclonal, #11303-1-AP, 1 to 150), METTL3 (rabbit polyclonal, #15073-1-AP, 1 to 1,500), or Ki67 (rabbit polyclonal, #27309-1-AP, 1 to 6,000). After HRP secondary detection (goat anti-rabbit, #ab6721, 1 to 1,000, Abcam, Cambridge, UK) and DAB visualization (Beyotime), we counterstained the slides with hematoxylin (Beyotime).
Quantitative polymerase chain reaction (PCR) for mRNA expression
With the BeyoMagTM RNA Kit (Beyotime) and Beyotime-formulated protocols, we prepared total RNA from harvested tissues and cultured cells. The PrimeScript RT Kit (TaKaRa, Dalian, China) was applied to reverse-transcribe 1 µg RNA. We then used a PCR machine (Rotorgene 6000, Qiagen, Hilden, Germany) for quantitative PCR with SYBR Premix ExTaq (TaKaRa) and specific primers for SERPINE2 [5'-AAGAAACGCACTTTCGTGGC-3'-forward (F), 5'-GTGTGGGATGATGGCAGACA-3'-reverse (R)] or METTL3 (5'-TTGTCTCCAACCTTCCGTAGT-3'-F, 5'-CCAGATCAGAGAGGTGGTGTAG-3'-R). We plotted relative expression as line fold change (2−ΔΔCt) after normalization to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5'-ACAACTTTGGTATCGTGGAAGG-3'-F, 5'-GCCATCACGCCACAGTTTC-3'-R).
Protein blot analysis
Immunoblots for SERPINE2 and METTL3 expression were conducted as described (20), using specific antibodies (Proteintech) recognizing SERPINE2 (rabbit polyclonal, #11303-1-AP, 1 to 800) and METTL3 (rabbit polyclonal, #15073-1-AP, 1 to 1,500). For protein preparation from harvested tissues and cultured cells, we applied a Protein Co-extraction Kit as suggested by the producer (Beyotime). Protein extracts were electrophoresed on 10–12% sodium dodecyl sulfate (SDS) polyacrylamide gels. We then blotted the resulting gels onto Clear Blot membrane-p (ATTO, Tokyo, Japan). After primary antibody and horseradish peroxidase (HRP) secondary detection (1 to 8,000), we developed the signals with Excellent Chemiluminescent Substrate (ECL) Detection Kit (Invitrogen). The expression of β-actin (rabbit recombinant, #81115-1-RR, 1 to 8,000) was used as a loading control.
Cell proliferation, migration and invasion assays
As described by the vendors, we utilized the Click-iT 5-ethynyl-2'-deoxyuridine (EdU)-488 Cell Proliferation Kit (Servicebio, Wuhan, China) and 4',6-diamidino-2-phenylindole (DAPI) staining (Beyotime) for evaluation of proliferative ability of GSRCC cells after the relevant transfection. We performed in vitro invasion and migration assays with the aid of Matrigel invasion chambers (BD Biosciences, Stockholm, Sweden) and 24-Transwells (BD Biosciences) as described elsewhere (21). Briefly, we plated serum-starved GSRCC cells after transfection into each upper chamber, and complete media were added into the lower chambers. After the GSRCC cells were allowed to invade or migrate for 24 h, we quantitated invaded or migratory cells upon crystal violet (1%, Beyotime) staining under the Olympus X-71 microscope (Olympus, Tokyo, Japan).
Tube formation assay
For tube formation assay, we seeded HUVECs (2×104 cells/well) re-suspended in 200 µL of the CM from transfected NUGC-4 and SNU-668 cells into 24-well plates precoated with Matrigel (BD Biosciences). After that, the incubation was allowed for 12 h. Under the Olympus microscope, we determined HUVEC tube formation ability by analyzing the number of tube junctions.
Flow cytometry for evaluation of apoptosis and M2-polarized macrophages
For FACS analyses, we used an LSRII cytometer (BD Biosciences) along with FlowJo 9.1 software (TreeStar, Ashland, OR, USA). Apoptosis of transfected NUGC-4 and SNU-668 cells was monitored by Annexin V-FITC and PI staining in accordance with the Detection Kit and accompanying instructions (Beyotime). For analysis of influence on M2 macrophage polarization, we incubated THP-1-derived macrophages (THP-1/M0) with the CM from transfected NUGC-4 and SNU-668 cells for 24 h, followed by staining with PE/Cy7-labeled CD206 antibody (#ab270682, Abcam) and FITC-labeled CD11b antibody (#FITC-65055, Proteintech).
Prediction of m6A sites of SERPINE2 mRNA and the METTL3-SERPINE2 relationship
We used the previously developed prediction server SRAMP (http://www.cuilab.cn/sramp) to predict the m6A methylation sites of SERPINE2 mRNA. By RBPsuite web (http://www.csbio.sjtu.edu.cn/bioinf/RBPsuite/), the potential binding of METTL3 with SERPINE2 mRNA was predicted.
Luciferase assays
For these assays, we first generated SERPINE2 reporter constructs by inserting SERPINE2 mRNA segment harboring the predicted m6A methylation site (WT-SERPINE2) or mutation in the target site (MUT-SERPINE2) into psiCHECK-2 vector (Promega, Paris, France). We then co-transfected into 293T cells (#C6008, Beyotime) with WT-SERPINE2 or MUT-SERPINE2 and sh-METTL3 or sh-NC. Lysates were prepared after 48 h. We assessed luciferase activity with Dual-Luciferase reporter system (Beyotime).
RNA immunoprecipitation (RIP) and methylated RIP (MeRIP) assays
We performed RIP and MeRIP experiments as described (19), using specific antibodies recognizing m6A (mouse monoclonal, #68055-1-Ig, 1 to 5,000, Proteintech), METTL3 (rabbit polyclonal, #15073-1-AP, 1 to 2,000, Proteintech), and immunoglobulin G (IgG) control (rabbit monoclonal, #ab172730, 1 to 1,000, Abcam). Briefly, lysates were prepared from NUGC-4 and SNU-668 cells with the BeyoRIP™ RIP Assay Kit (Beyotime), followed by incubation with the relevant antibody and protein A/G agarose (Beyotime) as suggested by the vendor. We harvested the beads and isolated m6A- or METTL3-associating RNA for evaluation of SERPINE2 mRNA enrichment.
Analysis of SERPINE2 mRNA stabilization
NUGC-4 and SNU-668 cells after 48 h transfection of sh-METTL3 or sh-NC were subjected to Actinomycin D (50 µg/mL, Beyotime) processing for 0, 4 and 8 h. We prepared RNA from these treated cells for assessment of SERPINE2 mRNA levels.
Statistical analysis
Data shown were means ± standard deviation (SD) of at least replicate experiments (n≥3). Comparisons of two groups were done using a Student’s t-test, and for multiple group comparisons, we used a two-way analysis of variance (ANOVA), with a probability value of less than 0.05 regarded statistically significant. We performed expression association analysis using Pearson’s correlation test.
Results
SERPINE2 expression is upregulated in human GSRCC
Using a previously developed detection algorithm GSE33335 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33335), we analyzed DEGs of a cohort of gastric tumors and matched normal gastric tissues. As illustrated in Figure 1A,1B, volcano plots showed many DEGs (P<0.05, |log2foldchange| ≥1) in adenocarcinoma, partial GSRCC samples versus paracancerous normal gastric tissues or adenocarcinoma tumors. Through combination of these DEGs (GSRCC vs. normal and GSRCC vs. adenocarcinoma), we found five related genes (CXCL5, SERPINE2, FGL2, GZMA, and ADGRG7), as shown in Venn diagram (Figure 1C). Since GSRCC is a diffuse type of gastric carcinoma (1), we used the KM-plotter algorithm (http://kmplot.com/analysis/index.php?p=service) to further examine the association of the five genes with prognosis of GSRCC by using diffuse gastric cancer samples (n=241). Among the five genes, only SERPINE2 was observed to negatively correlate with overall survival of patients with diffuse gastric cancer, and high levels of SERPINE2 foreboded a worse prognosis (Figure 1D and Figure S1). The distinct characteristic of SERPINE2 in GSRCC and its function in influencing gastric carcinogenesis (11,12) led us to study SERPINE2 expression, activity and regulation in GSRCC.
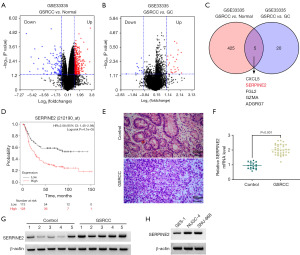
To confirm these bioinformatics findings, we performed a more quantitative measurement of SERPINE2 expression in GSRCC samples H&E staining revealed significant histological differences in GSRCC samples compared to normal gastric tissues. Normal gastric tissue typically showed well-organized glandular structures with uniform cell morphology, and GSRCC tumor cells were often diffusely scattered (Figure 1E). Consistent with previous findings (11,12), SERPINE2 mRNA and protein levels were upregulated in GSRCC samples relative to paracancerous normal gastric tissues (Figure 1F,1G). Similarly, high expression of SERPINE2 was observed in two GSRCC cell lines (NUGC-4 and SNU-668) versus normal GES-1 cells (Figure 1H).
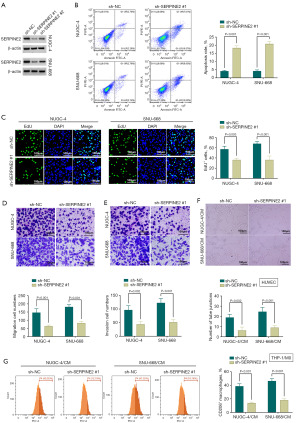
SERPINE2 deficiency suppresses GSRCC cell malignant phenotypes and M2 macrophage polarization
Although the important implication of SERPINE2 in gastric carcinogenesis has been established (11,12), its precise activity in GSRCC development is yet to be examined. Having demonstrated the upregulation of SERPINE2 in GSRCC, we sought to elucidate the influence of its deficiency in NUGC-4 and SNU-668 GSRCC cells, which exhibited high levels of SERPINE2 (Figure 1H). To model SERPINE2 deficiency in GSRCC, we utilized SERPINE2-shRNAs (sh-SERPINE2#1 and sh-SERPINE2#2), which efficiently depleted SERPINE2 in NUGC-4 and SNU-668 cells (Figure 2A). Since sh-SERPINE2#1 had stronger depletion efficiency (Figure 2A), we used it in the subsequent experiments. SERPINE2 depletion significantly enhanced in vitro apoptosis (Figure 2B). Conversely, deficiency of SERPINE2 diminished the proliferative, migratory and invasive capacities of NUGC-4 and SNU-668 cells (Figure 2C-2E). Abnormal angiogenesis contributes to tumor immune evasion and progression, and anti-angiogenic agents have been proposed for cancer therapy (3). We then examined the impact of SERPINE2 depletion on tube formation ability of HUVECs. By incubation of HUVECs with the CM from SERPINE2-depleted GSRCC cells, we observed that SERPINE2-depleted CM led to a remarkable defect in HUVEC tube formation (Figure 2F), suggesting that SERPINE2 deficiency may suppress angiogenesis in GSRCC. M2-polarized cancer-associated macrophages are well-known contributors to pro-tumorigenic processes in gastric carcinoma (22). M2 polarization suppressors have gained great attention due to their anti-cancer properties (4). To test the influence of SERPINE2 in M2 polarization, we incubated THP-1/M0 cells with the SERPINE2-depleted CM and found that SERPINE2 deficiency reduced the percent of the CD206+ macrophages compared with the sh-NC control (Figure 2G). Collectively, these data demonstrate that SERPINE2 depletion is capable of hindering GSRCC cell malignant phenotypes, HUVEC tube formation and M2 macrophage polarization.
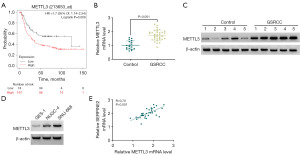
METTL3 is highly expressed in human GSRCC
As a methyltransferase, METTL3 has emerged as a vital regulator in a variety of cancers, including gastric carcinoma (17,18). When we used the KM-plotter algorithm to analyze the prognosis association of METTL3 with diffuse gastric cancer (n=241), we found that high METTL3 expression foreboded a poor prognosis (Figure 3A). Through expression analyses by quantitative PCR (qPCR) and immunoblot, in a cohort of fresh-frozen GSRCC samples, we observed increased levels of METTL3 mRNA and protein (Figure 3B,3C). Consistently, NUGC-4 and SNU-668 GSRCC cells had higher protein levels of METTL3 (Figure 3D). Furthermore, in GSRCC samples, expression association analysis unveiled the positive association of SERPINE2 and METTL3 (Figure 3E).
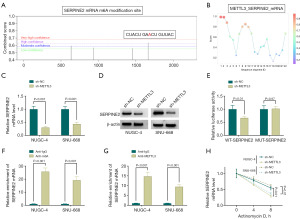
SERPINE2 is epigenetically modulated by METTL3 via m6A methylation modification
The METTL3 plays a vital role in gastric carcinoma depending on its regulation of RNA m6A methylation modification (17,23). Given the significantly positive association of SERPINE2 and METTL3 in GSRCC (Figure 3E), we hypothesized that METTL3 is likely to be responsible for SERPINE2 upregulation in GSRCC. Using the previously developed prediction server SRAMP (http://www.cuilab.cn/sramp), we found a putative m6A methylation site with very high confidence in SERPINE2 mRNA (Figure 4A). Moreover, the RBPsuite web (http://www.csbio.sjtu.edu.cn/bioinf/RBPsuite/) predicted the potential binding of METTL3 and SERPINE2 mRNA (Figure 4B). Moreover, in NUGC-4 and SNU-668 GSRCC cells, depletion of METTL3 reduced expression of SERPINE2 at both mRNA and protein (Figure 4C,4D). We then examined the influence of METTL3 in SERPINE2 mRNA by luciferase assays with wide-type (WT) SERPINE2 reporter construct carrying the putative m6A methylation site and mutant (MUT) construct in the target site. Disruption of METTL3 resulted in a striking reduction of luciferase activity of 293T cells transfected with WT-SERPINE2 construct, while it did not influence luciferase of MUT-SERPINE2 construct (Figure 4E). Our MeRIP-qPCR assays further showed the existence of m6A modified SERPINE2 mRNA in both NUGC-4 and SNU-668 cell lines, as evidenced by the abundant enrichment of SERPINE2 mRNA in the m6A-specific antibody precipitates (Figure 4F). Furthermore, RIP experiments with an anti-METTL3 antibody confirmed the binding of METTL3 with the SERPINE2 mRNA (Figure 4G). The regulation of METTL3 in SERPINE2 mRNA stabilization was also tested. Under actinomycin D conditions for transcription inhibition, depletion of METTL3 weakened the stability of SERPINE2 mRNA (Figure 4H). In sum, our findings support that SERPINE2 is epigenetically modulated by METTL3.
SERPINE2 is a mediator of METTL3 in regulating GSRCC cell malignant phenotypes and M2 macrophage polarization
Further, we sought to examine the effects of METTL3 silencing on cell phenotypes and the potential contribution of SERPINE2 to the effects of METTL3 silencing in NUGC-4 and SNU-668 GSRCC cells. In addition to the inhibition on SERPINE2 expression (Figure 5A), METTL3 silencing by sh-METTL3 introduction induced cell apoptosis (Figure 5B) and downregulated cell proliferative, migratory and invasive capacities (Figure 5C-5E). Moreover, via incubation with the CM of METTL3-silenced GSRCC cells, HUVECs displayed reduced tube formation ability (Figure 5F), and THP-1-derived macrophages exhibited lower percent of the CD206+ cells (Figure 5G,5H). These data indicate that, and consistent with the activity of SERPINE2 deficiency, depletion of METTL3 represses GSRCC cell malignant phenotypes, HUVEC tube formation and M2 macrophage polarization.
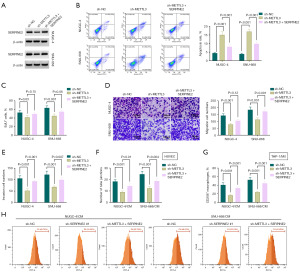
Based on the downregulation of SERPINE2 in METTL3-silenced GSRCC cells, we performed rescue experiments by restoring SERPINE2 expression. A recombinant SERPINE2 plasmid was used and confirmed to increase SERPINE2 protein levels in METTL3-silenced GSRCC cells (Figure 5A). Remarkably, restoration of SERPINE2 abolished the induction of apoptosis elicited by METTL3 silencing (Figure 5B). Additionally, restoration of SERPINE2 rescued METTL3 silencing-imposed defects of proliferative, migratory and invasive capacities (Figure 5C-5E). Furthermore, restored expression of SERPINE2 strikingly abrogated METTL3 silencing-imposed suppression of HUVEC tube formation (Figure 5F) and M2 macrophage polarization (Figure 5G,5H). Overall, our results suggest that the effects of METTL3 depletion on cell phenotypes are due to the reduction of SERPINE2 expression.
METTL3 deficiency inhibits xenograft growth in vivo
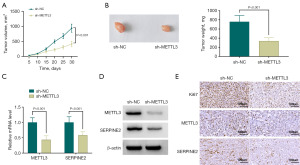
To test the influence of METTL3 depletion on tumorigenicity of GSRCC cells in vivo, we selected SNU-668 GSRCC cells and implanted the cells expressing sh-METTL3 subcutaneously in the nude mice (BALB/c). We allowed these xenograft tumors to grow for 30 days. Depletion of METTL3 had an inhibitory effect on xenograft growth (Figure 6A,6B). SNU-668 xenografts were further detected for expression of METTL3 and SERPINE2. We observed downregulated levels of METTL3 and SERPINE2 in xenografts formed by sh-METTL3-transduced SNU-668 cells (Figure 6C-6E). Finally, IHC data showed that the number of the Ki67-positive cells was diminished in sh-METTL3-transduced SNU-668 xenografts (Figure 6E), supporting the inhibitory impact of METTL3 deficiency on tumorigenicity of SNU-668 GSRCC cells.
Discussion
Critical molecular regulators responsible for the pro-tumorigenic process are under intensive exploration at present (24,25). Uncovering their activity and regulation will provide encouraging opportunities for targeted therapies against cancer. Our study reveals SERPINE2 as a crucial driver of GSRCC malignant progression. Dysregulation of RNA m6A methylation has the pro-tumorigenic activity in gastric carcinoma (15,16). Furthermore, we demonstrate that SERPINE2 is epigenetically modulated by METTL3, a critical regulator in gastric carcinoma progression (17,23), via m6A methylation modification, pointing to a novel epigenetic mechanism underlying GSRCC pathogenesis.
Aberrant SERPINE2 expression can contribute to oncogenesis and cancer metastasis in many aggressive malignancies, such as lung cancer, advanced renal cell carcinoma and liver cancer (6,10,26). High SERPINE2 expression is observed in gastric carcinoma samples and cell lines (11), which is consistent with our findings that SERPINE2 is upregulated in human GSRCC. Previous work also reveals the upregulation of SERPINE2 levels in advanced gastric carcinoma with aggressive lymph node metastasis (27), and high levels of SERPINE2 positively correlate with distant metastasis and lymph node metastasis (11). Our bioinformatics analysis (GSE33335 dataset) also showed the increased expression of SERPINE2 in GSRCC samples versus gastric adenocarcinoma tumors. These data suggest the association of high SERPINE2 expression with more aggressive gastric carcinoma and more advanced clinical stage. Our working model indicates that deficiency of SERPINE2 suppresses GSRCC cell malignant phenotypes and induces their apoptosis. Abnormal angiogenesis can induce cancer immune evasion and progression (3,28), and M2-polarized cancer-related macrophages promote the pro-tumorigenic processes in gastric cancer (4). By combining in vitro tube formation and M2 macrophage polarization analyses, we demonstrate the suppressive influence of SERPINE2 deficiency on GSRCC progression. Thus, SERPINE2 is likely to be a pro-tumorigenic driver of GSRCC. SERPINE2 has also been found to contribute to immunotherapeutic resistance by forming immunosuppressive microenvironment in gastric carcinoma (12). With these observations, we propose that silencing SERPINE2 may be a potential approach for GSRCC treatment.
The m6A writer METTL3, the sole catalytic subunit of the methyltransferase complex, actively takes part in a broad spectrum of biological processes due to its m6A RNA methyltransferase activity. Many documents prove that METTL3 epigenetically affects human carcinogenesis in a variety of cancer types (29). As an example, METTL3 enhances the stability of E2F1 mRNA via m6A modification to upregulate E2F1, thereby leading to colorectal cancer progression (30). On the other hand, although METTL3 induces m6A modification of the tumor inhibitor LATS1 mRNA, the concomitant m6A reader YTHDF2 recognizes m6A sites of LATS1 mRNA to diminish its stabilization, thus contributing to breast tumorigenesis (31). These reports suggest that METTL3-mediated mRNA stabilization or degradation through RNA m6A methylation may depend on the function of its concomitant m6A readers. In this report, we demonstrate that METTL3 induces SERPINE2 upregulation in GSRCC cells by enhancing SERPINE2 mRNA stabilization for the first time. In gastric carcinoma, METTL3 has been identified as a pro-tumorigenic driver by mediating m6A methylation of various mRNA transcripts, such as STAT5A and MIB1 (17,32). Moreover, METTL3 can accelerate angiogenesis and thus drives gastric tumorigenesis (33). Our data also indicate high expression of METTL3 in GSRCC samples and cell lines, implying the critical role of METTL3 in GSRCC progression. Furthermore, we demonstrate that depletion of METTL3 represses GSRCC cell in vitro malignant phenotypes, HUVEC tube formation and M2 macrophage polarization through SERPINE2 reduction, suggesting the pro-tumorigenic activity of the METTL3/SERPINE2 axis in GSRCC. Future work investigating the related m6A readers and the intracellular and extracellular signaling pathways related to GSRCC progression is warranted to expand our understanding of the epigenetic mechanism in GSRCC. In addition, our in vivo data find that METTL3 inhibition acts for xenograft growth suppression. Expanded analyses examining the potential contribution of SERPINE2 to the in vivo function of METTL3 are also warranted.
Conclusions
In summary, our work, revealing the METTL3/SERPINE2 axis as an epigenetic mechanism in GSRCC, highlights the significant roles of epigenetic dysregulation in GSRCC progression. Our observations may have diagnostic and/or therapeutic applications in GSRCC or other gastric carcinoma types with the upregulation of the METTL3/SERPINE2 axis.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-896/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-896/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-896/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-896/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Nuclear Industry 215 Hospital, Hospital of Shaanxi Province Ethics Committee (IRB No. 2023S29-UR1) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 6NI02-2023) granted by the Nuclear Industry 215 Hospital, Hospital of Shaanxi Province Animal Care and Use Committee, in compliance with national standard of the care and use of laboratory animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jiang Y, Hu H, Shao X, et al. A novel web-based dynamic prognostic nomogram for gastric signet ring cell carcinoma: a multicenter population-based study. Front Immunol 2024;15:1365834. [Crossref] [PubMed]
- Moradi L, Tajik F, Saeednejad Zanjani L, et al. Clinical significance of CD166 and HER-2 in different types of gastric cancer. Clin Transl Oncol 2024;26:664-81. [Crossref] [PubMed]
- Albini A, Noonan DM, Corradino P, et al. The Past and Future of Angiogenesis as a Target for Cancer Therapy and Prevention. Cancer Prev Res (Phila) 2024;17:289-303. [Crossref] [PubMed]
- Deng C, Huo M, Chu H, et al. Exosome circATP8A1 induces macrophage M2 polarization by regulating the miR-1-3p/STAT6 axis to promote gastric cancer progression. Mol Cancer 2024;23:49. [Crossref] [PubMed]
- Rosendal E, Mihai IS, Becker M, et al. Serine Protease Inhibitors Restrict Host Susceptibility to SARS-CoV-2 Infections. mBio 2022;13:e0089222. [Crossref] [PubMed]
- Zhang S, Jia X, Dai H, et al. SERPINE2 promotes liver cancer metastasis by inhibiting c-Cbl-mediated EGFR ubiquitination and degradation. Cancer Commun (Lond) 2024;44:384-407. [Crossref] [PubMed]
- Qu H, Jiang J, Zhan X, et al. Integrating artificial intelligence in osteosarcoma prognosis: the prognostic significance of SERPINE2 and CPT1B biomarkers. Sci Rep 2024;14:4318. [Crossref] [PubMed]
- Liu Y, Li X, Chen S, et al. Pan-cancer analysis of SERPINE family genes as biomarkers of cancer prognosis and response to therapy. Front Mol Biosci 2023;10:1277508. [Crossref] [PubMed]
- Chuang HW, Lin LH, Ji DD, et al. Serpin peptidase inhibitor, clade E, member 2 is associated with malignant progression and clinical prognosis in oral squamous cell carcinoma. J Dent Sci 2024;19:70-8. [Crossref] [PubMed]
- Chen WJ, Dong KQ, Pan XW, et al. Single-cell RNA-seq integrated with multi-omics reveals SERPINE2 as a target for metastasis in advanced renal cell carcinoma. Cell Death Dis 2023;14:30. [Crossref] [PubMed]
- Wang K, Wang B, Xing AY, et al. Prognostic significance of SERPINE2 in gastric cancer and its biological function in SGC7901 cells. J Cancer Res Clin Oncol 2015;141:805-12. [Crossref] [PubMed]
- Zhang D, Sun R, Di C, et al. Microdissection of cancer-associated fibroblast infiltration subtypes unveils the secreted SERPINE2 contributing to immunosuppressive microenvironment and immuotherapeutic resistance in gastric cancer: A large-scale study integrating bulk and single-cell transcriptome profiling. Comput Biol Med 2023;166:107406. [Crossref] [PubMed]
- Christodoulidis G, Koumarelas KE, Kouliou MN, et al. Gastric Cancer in the Era of Epigenetics. Int J Mol Sci 2024;25:3381. [Crossref] [PubMed]
- Li Y, Jin H, Li Q, et al. The role of RNA methylation in tumor immunity and its potential in immunotherapy. Mol Cancer 2024;23:130. [Crossref] [PubMed]
- Wu S, Li C, Zhou H, et al. The regulatory mechanism of m6A modification in gastric cancer. Discov Oncol 2024;15:283. [Crossref] [PubMed]
- Jiang T, Xia Y, Li Y, et al. TRIM29 promotes antitumor immunity through enhancing IGF2BP1 ubiquitination and subsequent PD-L1 downregulation in gastric cancer. Cancer Lett 2024;581:216510. [Crossref] [PubMed]
- Zang Y, Tian Z, Wang D, et al. METTL3-mediated N(6)-methyladenosine modification of STAT5A promotes gastric cancer progression by regulating KLF4. Oncogene 2024;43:2338-54. [Crossref] [PubMed]
- Gu C, Xia Y, Lu C, et al. TRIM50 inhibits glycolysis and the malignant progression of gastric cancer by ubiquitinating PGK1. Int J Biol Sci 2024;20:3656-74. [Crossref] [PubMed]
- Hu Z, Chen G, Zhao Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer 2023;22:55. [Crossref] [PubMed]
- Sun Y, Shen W, Hu S, et al. METTL3 promotes chemoresistance in small cell lung cancer by inducing mitophagy. J Exp Clin Cancer Res 2023;42:65. [Crossref] [PubMed]
- Liu CH, Zhang JJ, Zhang QJ, et al. METTL3 regulates the proliferation, metastasis and EMT progression of bladder cancer through P3H4. Cell Signal 2024;113:110971. [Crossref] [PubMed]
- Wang Y, Zhang J, Shi H, et al. M2 Tumor-Associated Macrophages-Derived Exosomal MALAT1 Promotes Glycolysis and Gastric Cancer Progression. Adv Sci (Weinh) 2024;11:e2309298. [Crossref] [PubMed]
- Liu T, Feng YL, Wang RY, et al. Long-term MNNG exposure promotes gastric carcinogenesis by activating METTL3/m6A/miR1184 axis-mediated epithelial-mesenchymal transition. Sci Total Environ 2024;913:169752. [Crossref] [PubMed]
- Liu Z, Zhou X, Tang F. Epigenetic regulators as the foundation for molecular classification of colorectal cancer. Cancer Biol Med 2024;21:547-52. [Crossref] [PubMed]
- Cai W, Xiao C, Fan T, et al. Targeting LSD1 in cancer: Molecular elucidation and recent advances. Cancer Lett 2024;598:217093. [Crossref] [PubMed]
- Zhang J, Wu Q, Zhu L, et al. SERPINE2/PN-1 regulates the DNA damage response and radioresistance by activating ATM in lung cancer. Cancer Lett 2022;524:268-83. [Crossref] [PubMed]
- Suh YS, Yu J, Kim BC, et al. Overexpression of Plasminogen Activator Inhibitor-1 in Advanced Gastric Cancer with Aggressive Lymph Node Metastasis. Cancer Res Treat 2015;47:718-26. [Crossref] [PubMed]
- Guo L, Xu B, Hu B, et al. Synthesis and Biological Evaluation of 4-substituted Aryl-piperazine Derivatives of 1,7,8,9-tetrachloro-10,10-dimethoxy-4-azatricyclo [5.2.1.02,6] dec-8-ene-3,5-dione as Anti-cancer and Anti-angiogenesis Agents. Letters in Drug Design & Discovery 2023;20:1538-43.
- Yu H, Liu J, Bu X, et al. Targeting METTL3 reprograms the tumor microenvironment to improve cancer immunotherapy. Cell Chem Biol 2024;31:776-791.e7. [Crossref] [PubMed]
- Chen Z, Zhou J, Wu Y, et al. METTL3 promotes cellular senescence of colorectal cancer via modulation of CDKN2B transcription and mRNA stability. Oncogene 2024;43:976-91. [Crossref] [PubMed]
- Xu Y, Song M, Hong Z, et al. The N6-methyladenosine METTL3 regulates tumorigenesis and glycolysis by mediating m6A methylation of the tumor suppressor LATS1 in breast cancer. J Exp Clin Cancer Res 2023;42:10. [Crossref] [PubMed]
- Xu P, Liu K, Huang S, et al. N(6)-methyladenosine-modified MIB1 promotes stemness properties and peritoneal metastasis of gastric cancer cells by ubiquitinating DDX3X. Gastric Cancer 2024;27:275-91. [Crossref] [PubMed]
- Wang N, Huo X, Zhang B, et al. METTL3-Mediated ADAMTS9 Suppression Facilitates Angiogenesis and Carcinogenesis in Gastric Cancer. Front Oncol 2022;12:861807. [Crossref] [PubMed]


