
LncRNA ZEB1-AS1 promotes the proliferation and migration of non-small cell lung cancer by activating epithelial-mesenchymal transition with STAT3
Highlight box
Key findings
• The overexpression of long non-coding RNA ZEB1 antisense 1 (lncRNA ZEB1-AS1) increased the protein levels of ZEB1 and STAT3, promoted the occurrence of epithelial-mesenchymal transition (EMT), and enhanced the invasion and migration abilities of lung cancer cells.
What is known and what is new?
• LncRNA ZEB1-AS1 promotes the expression of ZEB1 and cancer progression in bladder cancer.
• LncRNA ZEB1-AS1 is associated with a poor prognosis, promotes EMT via ZEB1 and STAT3 in non-small cell lung cancer (NSCLC cells), and up-regulates STAT3 expression by competitively binding to miRNA519d.
What is the implication, and what should change now?
• LncRNA ZEB1-AS1 is closely related to the EMT of NSCLC cells, and may be an important target for inhibiting the proliferation and metastasis of NSCLC. Determining the role of ZEB1-AS1 in the occurrence of NSCLC could not only improve the understanding of tumor metastasis caused by lncRNA, but could also contribute to the development of new treatment strategies for NSCLC.
Introduction
Lung cancer is the most common cause of cancer-related death worldwide, and causes approximately 1.6 million deaths annually (1). There are two main types of lung cancer: non-small cell lung cancer (NSCLC), which accounts for 85% of cases, and small cell lung cancer (SCLC), which accounts for 15% of cases (2). Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma are the two most common subtypes of NSCLC (3). The most common cause of lung cancer is smoking, but LUAD is also common in nonsmokers (4).
Epithelial-mesenchymal transition (EMT) is a complex process that commonly occurs in tumors. During this process, the epithelial cells lose their polarity and cell-to-cell connections, which in turn enhances their migration and invasion abilities (5). The transcriptional repressor ZEB1 regulates neuronal differentiation under normal physiological functions, but substantial research suggests that ZEB1 plays a critical role in driving EMT in tumor cells (6). Transcription factors, such as ZEB1, ZEB2, Snai1, and Twist1, are calcium-dependent transmembrane glycoproteins, and E-cadherin mediates the mutual adhesion of cells of the same species. E-cadherin is a calcium-dependent transmembrane glycoprotein. E-cadherin participate in the formation and maintenance of normal intercellular junctions, and is the most important class of protein molecules that mediate the intercellular junctions, the transcription factors ZEB1, ZEB2, Snai1, and Twist1 are its direct inhibitory factors (7,8).
Growing evidence has identified the link between dysregulation of long non-coding RNAs (lncRNAs) and microRNAs in cancer progression (9). LncRNA LINC00240 suppresses invasion and migration in NSCLC by sponging miR-7-5p (10). LncRNA SNHG7 promotes NSCLC progression and cisplatin resistance by inducing autophagic activity. These evidences suggest that lncRNAs play an important role in regulating NSCLC biology (11).
LncRNA ZEB1-AS1 is derived from the ZEB1 promoter region. Studies have shown that lncRNA ZEB1-AS1 promotes the expression of ZEB1 in bladder cancer and glioblastoma, and cancer progression, and is also associated with a poor prognosis (12-14). However, the biological functions and molecular mechanism of lncRNA ZEB1-AS1 in NSCLC migration remain unclear.
In this study, by overexpressing or silencing lncRNA ZEB1-AS1, we found that ZEB1-AS1 promoted the invasion and migration of lung cancer cells, and the overexpression of ZEB1-AS1 increased the protein levels of ZEB1 and STAT3, thus promoting the occurrence of EMT in lung cancer cells. Based on our in-depth analysis of this mechanism, we suggest that lncRNA ZEB1-AS1 could be used as a molecular sponge to form a competing endogenous RNA (ceRNA) regulatory network of lncRNA ZEB1-AS1~miRNA519d~STAT3, thereby promoting the expression of STAT3. Our findings indicate ZEB1-AS1 is closely related to EMT in NSCLC, and may be an important target for inhibiting the proliferation and migration of NSCLC. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2276/rc).
Methods
Cell culture
The human NSCLC cell lines A549 and H1650 were purchased from National Infrastructure of Cell Line Resource (A549: SCSP-503; H1650: SCSP-592). Both A549 and NCI-H1650 were cultured in Roswell Park Memorial Institute Medium-1640 (11875093, Thermo Fisher Scientific, Waltham, MA, USA), containing 10% fetal bovine serum (A5256701, Thermo Fisher Scientific), and 1% antibiotics (15140122, Thermo Fisher Scientific) was added to the complete medium. All the cells were incubated in a 5% carbon dioxide (CO2) environment at 37 ℃.
Cell transfection
LncRNA ZEB1-AS1 was cloned by rapid-amplification of cDNA ends (RACE); the primers are detailed in Table S1. A recombinant adenovirus with overexpressing lncRNA ZEB1-AS1 was constructed using the AdEasy system (Agilent, Santa Clara, CA, USA) (15). In brief, the lncRNA ZEB1-AS1 gene was cloned into the pAdTrack-CMV shuttle vector (16405, Addgene, Cambridge, MA, USA; the primers are detailed in Table S1), digested with Pme I, and linearized. Linearized pAdTrack-CMV was then co-transformed with the pAdEasy-1 backbone (16400, Addgene) into the BJ5183 cells. After kanamycin resistance selection, the recombinant plasmid was transfected into the 293-cell package, and the recombinant adenovirus was harvested after 10 days of culture. The recombinant adenovirus was then infected with the NSCLC cells. The small interfering RNA (siRNA) sequences were derived as described previously by Li et al. (16) (see Table S1). The siRNA was synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China), and used to transfect the lncRNA ZEB1-AS1 overexpressing cells at a final concentration of 100 nM, which instantly inhibited the expression of the ZEB1 gene.
Using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) as the reagent, the transient transfection was performed in accordance with the manufacturer’s instructions. In brief, seed cells to be 70–90% confluent a transfection; next, dilute Lipofectamine™ 3000 reagent in Opti-MEM™ Medium; dilute DNA in Opti-MEM™ medium, then add P3000™ reagent and mix well; add the diluted DNA obtained in the previous step to the diluted Lipofectamine™ 3000 reagent (1:1). The cells were incubated for 10–15 minutes at room temperature, and DNA-lipid complex was then added to the cells. The incubated cells were tested 24 hours after transfection. Micro RNA (MiR)-519d-3p mimic (micrON hsa-miR-519d-3p mimic, #miR10002853-1-5, Shanghai, China) and mimic negative control (micrON mimic NC, #miR1N0000001-1-10) were purchased from BIROBIO Co., Ltd. (Guangzhou, China); the sequences are detailed in Table S1. MiR-519 mimic and mimic NC were transfected into the cells using Lipofectamine™ RNAiMAX Reagent (13778075, Invitrogen) in accordance with the manufacturer’s instructions. The siZEB transfection successfully knocked down ZEB1 expression (Figure S1A). The miR-519 mimic transfection successfully increased miR-519 expression (Figure S1B).
Dual-luciferase assay
The three prime untranslated region (3'UTR) of STAT3, and lncRNA ZEB1-AS1 and its mutated sequences were cloned into the pEZX-MT01 vector (Genecopeia, Rockville, MD, USA). The human embryonic kidney cells (HEK293) were co-transfected with 0.1 µg of the pEZX-MT01 vector (a firefly luciferase reporter construct) and 0.01 µg of pRLTK Renilla luciferase. The miR-519 mimic or the mimic Ctrl were co-transfected using Lipofectamine™ 2000 (11668030, Invitrogen). The sequences are detailed in Table S2. The activities of the firefly and Renilla luciferases were measured 48 hours after co-transfection using the dual-luciferase assay kit (Promega, Madison, WI, USA) and a Glomax 96 microplate spectrophotometer (Promega).
Transwell assay
A Transwell Cell Staining Kit (BL710A, Biosharp, Hefei, China) was used to evaluate the effect of ZEB1-AS1 on the invasion ability of the NSCLC cells. In brief, the cells in the logarithmic growth phase were taken from different groups, and starved for 1 day. The cells were then washed three times with phosphate-buffered saline (PBS), and treated with 0.25% trypsin (25200056, Thermo Fisher Scientific). Next, the cells were collected by centrifugation at 1,000 rpm/min at 4 ℃. After washing for three times with PBS, the cells were added to 2 mL of serum-free medium. The required single-cell suspension was then prepared, and the cell concentration was adjusted to 2×105 cells/mL. Next, the cells were transferred to the wells that had been pre-coated with Matrigel and incubated at 37 ℃. The cells were taken out in groups from the chamber at 24 and 48 hours, and counted after staining with crystal violet. Finally, an inverted microscope was used for observation. Five observation fields were randomly chosen and photographed, and the number of cells passing through the membrane was recorded.
Wound healing analysis
The effect of ZEB1-AS1 on the migration ability of the NSCLC cells was evaluated using the cell scratch assay. In brief, 1 mL of cell suspension with a density of approximately 1×105 was inoculated into a six-well plate, and cultured for 24 hours; each group of cells had three replicates. After adherence, the culture medium was added to form the monolayer cells, and a 100-µL sterile tip was used to vertically scratch the monolayer cells. The detached cells were washed with PBS. The cells were then incubated in an incubator containing 5% CO2 at 37 ℃. The cells were observed and photographed at 0, 24, 48, and 72 hours, respectively. To reduce the influence of cell proliferation on the migration results, serum-free medium was used in the scratch experiment, and 1 µg/mL of mitomycin was used to treat the cells for 1 hour to inhibit cell division.
Real-time quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted using the TRIzol kit (15596026, Invitrogen). In accordance with the kit’s instructions, reverse transcription was performed on the total RNA using the GoScriptTM Reverse Transcription Mix (Promega) to obtain the complementary DNA (cDNA). In the RT-qPCR, the cDNA amplification was performed on the CFX96 Touch Real-Time System (Bio-Rad, Hercules, CA, USA) using the SYBR Premix Ex Taq kit (RR390A, TaKaRa, Dalian, China). The prepared PCR reaction solution contained the following components: SYBR Premix Ex TaqTM II (2×): 12.5 µL; PCR forward primer (10 µM): 1 µL; PCR reverse primer (10 µM): 1 µL; DNA template: 2 µL; and ddH2O: 8.5 µL. The melting curve steps of the thermal program were as follows: 30 s at 95 ℃; 40 cycles at 5 s at 95 ℃, 30 s at 60 ℃, and a gradual increase from 65 ℃ to 95 ℃ at 0.5 ℃ s−1; the data were collected every 6 s. The RT-qPCR data were analyzed using 2−ΔΔCt with β-Actin as the beta-actin. See Table 1 for the primer sequences.
Table 1
| Gene name | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| Snail | GTGTCTCCCAGAACTATT | GTTTGAAATATAAATACCAGTGT |
| STAT3 | AGAGATTGACCAGCAGTA | CTTGATTCTTCGTAGATTGTG |
| Twist | ACCATCCTCACACCTCTG | GATTGGCACGACCTCTTG |
| ZEB1 | CAGATGAAGCAGGATGTA | CAGTGTCTTGTTGTTGTAG |
| E-cadherin | GTCGGATTATGCTATTGA | TAGTCTGTCTATCGTGAA |
| Integrins | ATTGACCTCTACTACCTT | GTGTTGTGCTAATGTAAG |
| Vimentin | TCAGAATATGAAGGAGGAA | GGCAATCTCAATGTCAAG |
| β-Actin | CTCTTCCAGCCTTCCTTCCT | AGCACTGTGTTGGCGTACAG |
qPCR, quantitative polymerase chain reaction.
Western blot
The cells from different groups were quickly transferred to the pre-chilled RIPA lysis buffer containing protease inhibitors (11836170001, Roche, Basel, Switzerland) to extract the total protein. The protein separation was performed using 8% sodium dodecyl-sulfate polyacrylamide gel electrophoresis, and then transferred to the polyvinylidene fluoride membranes. The membranes were washed with Tris-Buffered Saline with Tween (TBST) for 10 min, and blocked in TBST containing 5% bovine serum albumin for 1 h at room temperature. Next, the membranes were incubated overnight using the primary antibodies (E-cadherin, GTX100443, GeneTex, San Antonio, TX, USA; N-cadherin, GTX127345 GeneTex; Vimentin, ab92547, Abcam, Cambridge, UK; Snail, ab216347, Abcam, UK; Twist, ab187008, Abcam, UK; integrins ab150361, Abcam, UK; STAT3, ab68153, Abcam, UK; MMP-9, ab76003, Abcam, UK; β-Actin, ab8227, Abcam, UK). After washing the membrane with TBST, the membranes were incubated using the secondary antibody (1:5,000, ab205718, Abcam, UK) for 2 hours. Next, the ECL kit (Millipore, Billerica, MA, USA) was used to perform the color reaction, and the ImageJ software (http://imagej.nih.gov/ij/) was used to conduct the grayscale analysis.
Radioimmunoprecipitation (RIP)
The cells were collected in the logarithmic growth phase, and subsequently, the nuclei were pelleted through centrifugation. The cell nuclei were then re-suspended in RIP buffer. The suspension was mechanically sheared and precipitated to obtain chromatin. The supernatant was incubated overnight at 4 ℃ with protein antibodies [anti-immunoglobulin G (IgG), ab131368; and anti-AUF1, ab259895, Abcam, Shanghai, China]. Protein A/G beads (ab286842, Abcam, Shanghai, China) were used to bind the proteins. Co-precipitated RNA was isolated using the TRIzol RNA extraction reagent (15596026CN, Invitrogen) in accordance with the manufacturer’s instructions. The reverse transcription process was performed as outlined in the RT-PCR section above.
Statistical analysis
SPSS 21.0 software (IBM, Armonk, NY, USA) was used for the statistical analysis. The least significant difference t-test (unpaired) and a one-way analysis of variance were used to compare the data. Tukey’s post-hoc test was used to analyze the datasets containing three groups. The Spearman test was used for the correlation analysis. A P value <0.05 indicated a statistically significant difference.
Results
ZEB1-AS1 overexpression promotes the invasion and migration of NSCLC cells
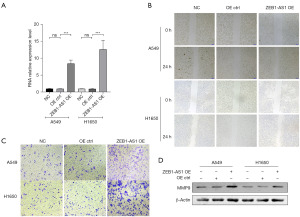
We established an overexpressed cell line of lncRNA ZEB1-AS1 using the lentiviral vector. RT-qPCR was performed to detect the expression level of lncRNA ZEB1-AS1 in the overexpressed cell line. The results show that compared with the cells in the overexpressed control (OE CTRL) group and the negative control (NC) group, the expression of lncRNA ZEB1-AS1 in the overexpressed cell line (ZEB1-AS1 OE) was significantly increased (P<0.001) (Figure 1A), which indicates that the overexpressed cell line had been successfully constructed. We then performed the wound healing assay and the Transwell invasion assay to determine the effect of ZEB1-AS1 overexpression on the migration and invasion abilities of the NSCLC cells. The results of the wound healing assay showed that compared with the NC and OE CTRL groups, the wound healing speed of the ZEB1-AS1 OE group was significantly enhanced, indicating that the overexpression of ZEB1-AS1 promoted the migration ability of NSCLC cells (Figure 1B). The Transwell assay showed that the number of transmembrane cells in the ZEB1-AS1 OE group was significantly higher than those in the NC and OE CTRL groups, indicating that the overexpression of ZEB1-AS1 increased the invasion ability of NSCLC cells (Figure 1C). Finally, we examined the expression of the MMP-9 protein, which plays an important role in tumor invasion and metastasis, in the ZEB1-AS1 overexpressed cell line by western blot. The results showed that the overexpression of ZEB1-AS1 increased the protein expression level of MMP-9 (Figure 1D).
ZEB1-AS1 overexpression promotes EMT of NSCLC cells
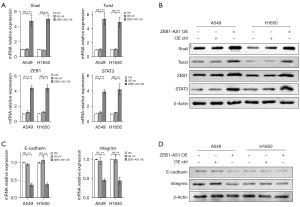
EMT is an important mechanism in the invasion and metastasis of primary tumors. Therefore, we examined the expression levels of EMT-related proteins after ZEB1-AS1 overexpression. First we detected the expression levels of Snail, Twist, ZEB1, and STAT3, which are the transcription factors most closely related to the occurrence of EMT. The RT-qPCR and western blot results showed that in the overexpressed ZEB1-AS1 cell line, the transcription and translation levels of these genes were all increased to varying degrees (Figure 2A,2B). Thus, the overexpression of ZEB1-AS1 significantly increased the expression of these transcription factors. In addition, after the overexpression of ZEB1-AS1, the expression of marker proteins Vimentin and N-cadherin were also up-regulated during the occurrence of EMT, while the expression levels of proteins E-cadherin and integrins were decreased (Figure 2C,2D). These results indicated that ZEB1-AS1 overexpression promoted the occurrence of EMT in the NSCLC cells.
ZEB1-AS1 participates in EMT via ZEB1 and STAT3
We knocked down ZEB1 in the ZEB1-AS1 overexpressed cell lines to show that ZEB1-AS1 promotes EMT in NSCLC by up-regulating ZEB1 expression. The western blot results of the expression levels of the EMT-related proteins showed that after the knockdown of ZEB1 in the ZEB1-AS1 overexpressed cell lines (OE + ZEB1 siRNA), the expression levels of Vimentin and N-cadherin were decreased, but their expression levels were still higher than the expression level of the control group of lung cancer cells (Figure 3A). Figure S2 shows the results of the quantitative analysis. The results showed that in the overexpressed ZEB1-AS1 NSCLC cell lines, the migration and invasion abilities of the cells were reduced after the knock down of ZEB1 (OE + ZEB1 siRNA), but they were still higher than those of the NSCLC cells in the control group, and this was particularly evident in the A549 cell line (Figure 3B,3C). Thus, we speculate that ZEB1 is not the only way by which ZEB1-AS1 participates in EMT.
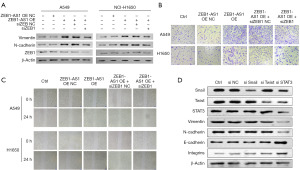
To further explore the possible mechanism by which ZEB1-AS1 participates in the EMT of NSCLC, we sought to knock down the transcription factors closely related to EMT in the ZEB1-AS1 overexpressed cell lines. The western blot results showed that when Snail and Twist were knocked down in the ZEB1-AS1 overexpressed cell lines, only a few EMT-related marker protein molecules changed. However, when STAT3 was knocked down in the ZEB1-AS1 overexpressed cell lines, the protein expression levels of Vimentin and N-cadherin were significantly decreased, while the protein levels of E-cadherin and the integrins were increased (Figure 3D). Figure S3 shows the results of the quantitative analysis. Therefore, ZEB1 and STAT3 appear to be the key factors by which ZEB1-AS1 participates in the regulation of EMT in tumor cells.
ZEB1-AS1~miRNA519d~STAT3 ceRNA network
In bladder cancer, ZEB1-AS1 promotes the translation of STAT3 messenger RNA (mRNA) by interacting with AUF1. Therefore, we used the RIP assay to determine whether the increase of the STAT3 protein level caused by ZEB1-AS1 overexpression was based on the promotion of STAT3 mRNA translation by AUF1. The results showed that both lncRNA ZEB1-AS1 and ZEB1 mRNA bind to AUF1, but no binding between AUF1 and STAT3 mRNA was detected (Figure 4A). According to a further bioinformatics analysis, both lncRNA ZEB1-AS1 and the STAT3 3'-UTRs bind to miRNA519d (Figure 4B); the corresponding sequences are provided in Table 2. Therefore, we used the dual-luciferase assay to carry out the further verification, and found that STAT3 was the target gene of miRNA519d, and lncRNA ZEB1-AS1 also binds to miRNA519d (Figure 4C). Compared with the control cells, after silencing ZEB1 and overexpressing miRNA519d in the lncRNA ZEB1-AS1 overexpressed cell lines, the mRNA and protein expression levels of lncRNA ZEB1-AS1 and STAT3 were down-regulated, while the mRNA and protein expression levels of the EMT protein markers Vimentin and N-cadherin were significantly reduced, and the mRNA and protein levels of E-cadherin and the integrins were increased (Figure 4D,4E).
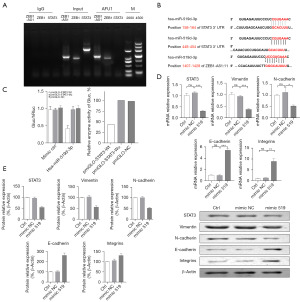
Table 2
| Sequence name | Sequence |
|---|---|
| hsa-miR-519d-3p | 3'-GUGAGAUUUCCCUCCGUGAAAC-5' |
| Position 158-164 of STAT3 3'UTR | 5'-CUUUGAGCAAUCUGGGCACUUUU-3' |
| Position 448-454 of STAT3 3'UTR | 5'-CAUACUCCUGGCAUUGCACUUUU-3' |
| Position 1407-1428 of ZEB1-AS1:11 | 5'-CCACAUUUUCTTGGGCAUUUUG-3' |
Discussion
Lung cancer is one of the most common malignant tumors worldwide, and NSCLC is the most important subtype of lung cancer. The most common cause of lung cancer is smoking, and eliminating the use of all tobacco-related products is a critical part of the global endeavor to prevent and treat lung cancer; however, research has shown that lung cancer is becoming increasingly common among women who have never smoked and in East Asia (17-20). Thus, it is of great importance that new molecular targets for lung cancer are found for the diagnosis, treatment, and prognosis of lung cancer.
Many previous studies have shown that the transcription factor ZEB1 plays an important role in both physiology and oncology (9,10). As a well-known promoter of EMT, ZEB1 plays a major role in cancer metastasis. According to a recent study, the antisense transcript lncRNA ZEB1-AS1 derived from the ZEB1 promoter positively regulates the expression of ZEB1 in hepatocellular carcinoma, and promotes cell growth and migration (16).
In this study, after constructing the ZEB1-AS1 overexpressing cells using the lentiviral vector, the invasion and migration abilities of the human NSCLC cell lines were both enhanced, and the protein expression level of MMP-9 was increased. MMP-9 is one of the most widely studied matrix metalloproteinases (MMPs), which cleaves many extracellular matrix (ECM) proteins to regulate the ECM remodeling. It also cleaves many plasma surface proteins, releasing them from the cell surfaces. MMP-9 has been found to be associated with multiple pathologies of cancer, including invasion, metastasis and angiogenesis (21). Based on the evaluation of many sample sets, Blanco-Prieto et al. (22) found that MMP-9 was present at higher levels in the serum of NSCLC patients compared with the healthy controls, suggesting that MMP-9 is a potential biomarker of NSCLC. In addition to the increased protein expression and enzymatic activity of MMP-9, the expression levels of the transcription factors snail, Twist, ZEB1, and STAT3, which are closely related to EMT, were also increased in the ZEB1-AS1 overexpressed cells to varying degrees. Moreover, the expression levels of the EMT marker proteins Vimentin and N-cadherin were also up-regulated after ZEB1-AS1 overexpression. These results indicate that the overexpression of ZEB1-AS1 promotes the occurrence of NSCLC EMT.
According to the study of Zhao et al. (13), ZEB1-AS1 promotes the translation of ZEB1 by recruiting AUF1 to the ZEB1 mRNA 3'UTR in bladder cancer cells. However, when we knocked down ZEB1 in the ZEB1-AS1 overexpressed cell lines, the expression levels of Vimentin and N-cadherin were decreased, but their expression levels were still higher than those in the lung cancer cells of the control group. Therefore, it can be deduced that ZEB1 is not the only way by which ZEB1-AS1 participates in EMT. To further explore the other mechanisms by which ZEB1-AS1 participates in NSCLC EMT, we knocked down the EMT-related transcription factors in the ZEB1-AS1 overexpressed cell lines. When ZEB1 and STAT3 were knocked down in the ZEB1-AS1 overexpressed cell lines, the protein expression levels of Vimentin and N-cadherin were significantly reduced. The above results suggest that the overexpression of ZEB1-AS1 promotes the occurrence of NSCLC EMT through ZEB1 and STAT3.
Based on the study of Zhao et al. (13), we assumed that the increased STAT3 protein levels caused by the ZEB1-AS1 overexpression was also based on the promotion of STAT3 mRNA translation by AUF1, and performed a RIP assay for verification. We found that AUF1 failed to bind with STAT3 mRNA. According to a further bioinformatics analysis, both ZEB1-AS1 and the STAT3 3'-UTR bind to miRNA519d. Thus, miRNA519d may participate in the ZEB1-AS1-STAT3-ZEB1 network as a regulatory mediator. The verification results of the dual-luciferase assay showed that STAT3 was the target gene of miRNA519d, and lncRNA ZEB1-AS1 also binds to miRNA519d, which provides further evidence in support of our hypothesis. However, our study still had some limitations, and we did not conduct animal experiments to further validate our conclusions. We intend to conduct follow-up animal experiments to gather further evidence in support of our conclusions in the future.
Conclusions
In conclusion, our study showed that lncRNA ZEB1-AS1 forms the ceRNA regulatory network of lncRNA ZEB1-AS1~miRNA519d~STAT3 as the molecular sponge, thereby promoting the expression of STAT3. ZEB1-AS1 is closely related to the EMT of NSCLC, which is an important target for inhibiting the proliferation and metastasis of NSCLC. Determining the role of ZEB1-AS1 in the occurrence of NSCLC could not only improve the understanding of tumor metastasis caused by lncRNA, but could also contribute to the development of new treatment strategies for NSCLC.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2276/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2276/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2276/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2276/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- La Salvia A, Meyer ML, Hirsch FR, et al. Rediscovering immunohistochemistry in lung cancer. Crit Rev Oncol Hematol 2024;200:104401. [Crossref] [PubMed]
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584-94. [Crossref] [PubMed]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer 2007;7:778-90. [Crossref] [PubMed]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013;13:97-110. [Crossref] [PubMed]
- Schwab A, Rao Z, Zhang J, et al. Zeb1 mediates EMT/plasticity-associated ferroptosis sensitivity in cancer cells by regulating lipogenic enzyme expression and phospholipid composition. Nat Cell Biol 2024;26:1470-81. [Crossref] [PubMed]
- Guo F, Parker Kerrigan BC, Yang D, et al. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. J Hematol Oncol 2014;7:19. [Crossref] [PubMed]
- Wang D, Du G, Chen X, et al. Zeb1-controlled metabolic plasticity enables remodeling of chromatin accessibility in the development of neuroendocrine prostate cancer. Cell Death Differ 2024;31:779-91. [Crossref] [PubMed]
- Gupta S, Silveira DA, Piedade GPS, et al. A dynamic Boolean network reveals that the BMI1 and MALAT1 axis is associated with drug resistance by limiting miR-145-5p in non-small cell lung cancer. Noncoding RNA Res 2024;9:185-93. [Crossref] [PubMed]
- Ku GW, Kang Y, Yu SL, et al. LncRNA LINC00240 suppresses invasion and migration in non-small cell lung cancer by sponging miR-7-5p. BMC Cancer 2021;21:44. [Crossref] [PubMed]
- She K, He S, Lu X, et al. LncRNA SNHG7 promotes non-small cell lung cancer progression and cisplatin resistance by inducing autophagic activity. J Thorac Dis 2023;15:155-67. [Crossref] [PubMed]
- Lin J, Zhan Y, Liu Y, et al. Increased expression of ZEB1-AS1 correlates with higher histopathological grade and promotes tumorigenesis in bladder cancer. Oncotarget 2017;8:24202-12. [Crossref] [PubMed]
- Zhao X, Wang D, Ding Y, et al. lncRNA ZEB1-AS1 promotes migration and metastasis of bladder cancer cells by post-transcriptional activation of ZEB1. Int J Mol Med 2019;44:196-206. [Crossref] [PubMed]
- Wang R, Xu F, Yang Z, et al. The mechanism of PFK-1 in the occurrence and development of bladder cancer by regulating ZEB1 lactylation. BMC Urol 2024;24:59. [Crossref] [PubMed]
- He TC, Zhou S, da Costa LT, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A 1998;95:2509-14. [Crossref] [PubMed]
- Li T, Xie J, Shen C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene 2016;35:1575-84. [Crossref] [PubMed]
- Alberg AJ, Brock MV, Ford JG, et al. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e1S-e29S.
- Vineis P, Airoldi L, Veglia F, et al. Environmental tobacco smoke and risk of respiratory cancer and chronic obstructive pulmonary disease in former smokers and never smokers in the EPIC prospective study. BMJ 2005;330:277. [Crossref] [PubMed]
- Hackshaw AK, Law MR, Wald NJ. The accumulated evidence on lung cancer and environmental tobacco smoke. BMJ 1997;315:980-8. [Crossref] [PubMed]
- Liao S, Wang Y, Zhou J, et al. Associations between chronic obstructive pulmonary disease and ten common cancers: novel insights from Mendelian randomization analyses. BMC Cancer 2024;24:601. [Crossref] [PubMed]
- Huang H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sensors (Basel) 2018;18:3249. [Crossref] [PubMed]
- Blanco-Prieto S, Barcia-Castro L, Páez de la Cadena M, et al. Relevance of matrix metalloproteases in non-small cell lung cancer diagnosis. BMC Cancer 2017;17:823. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


