
CD69 predicts prognosis through immune cell infiltration and decitabine treatment response in acute myeloid leukemia
Highlight box
Key findings
• CD69 expression was inversely related to overall survival (OS) of acute myeloid leukemia (AML) patients. A higher expression of CD69 was associated with reduced infiltration of CD8+ T cells and macrophages in AML. AML patients treated with decitabine (DA) exhibited decreased CD69 expression. The multifaceted role of CD69 in AML, encompassing its association with prognosis, immune cell infiltration, and response to chemotherapy, underscores its potential as a key player in AML pathogenesis and treatment response.
What is known and what is new?
• The tumor microenvironment (TME) has great impact on AML, and CD69 is a potential TME regulator. The precise relationship between CD69 and AML is yet to be fully elucidated.
• CD69 expression was inversely related to OS of AML patients. A higher expression of CD69 was associated with reduced infiltration of CD8+ T cells and macrophages in AML. AML patients treated with DA exhibited decreased CD69 expression.
What is the implication, and what should change now?
• The study established CD69 as a critical biomarker and molecular target in AML progression and immune cell infiltration. Based on our results, CD69 merits further research for a deeper understanding of its role in AML progression and immune regulation.
Introduction
Background
Acute myeloid leukemia (AML) is a lethal myeloid neoplasm characterized by unrestrained proliferation of hematopoietic stem cells accompanied by cytogenetic and molecular aberrations (1,2). It is highly heterogeneous with a generally poor prognosis and a high relapse rate.
Cytogenetic and molecular aberrations are pivotal in determining the response to chemotherapy and the long-term outcome in AML. For example, upregulation of stearoyl-CoA desaturase (SCD) in AML cells impedes ferroptosis and promotes proliferation (3). Bioinformatic analysis has highlighted SCD as a risk-associated gene and the SCD inhibitor A939572 has shown promising effects in the treatment of AML (4). Additionally, the S100 proteins, S100A8 and S100A9, have been extensively studied for their role in tumor signaling pathways. In AML, S100A8 accelerates tumor proliferation, while S100A9 modulates the toll-like receptor 4 (TLR4)-mitogen-activated protein kinase (MAPK) pathways, influencing cellular differentiation and survival (5,6). These insights underscore the necessity of further molecular research in AML for better prognostication and treatment strategies.
Alterations in the immune microenvironment are increasingly recognized as factors that exacerbate AML progression. Immunotherapy has shown significant antileukemia activity and N6-methyladenosine (m6A)-related long non-coding RNAs (lncRNAs)-based risk prediction models have been utilized to predict prognosis and design immunotherapy in AML patients (7). An investigation utilizing NanoString, bulk ribonucleic acid (RNA)-Seq and single-cell RNA-Seq data from independent clinical cohorts comprising 1,896 patients treated with chemotherapy and/or immune checkpoint blockade (ICB) demonstrated that new immune effector dysfunction (IED) scores provided improved AML-risk stratification. The IED expression signatures identified an ICB-unresponsive tumor microenvironment (TME) and predicted significantly shorter overall survival (OS) (8). Additionally, flotetuzumab, a bispecific antibody-based molecule targeting CD3ε and CD123 in a dual affinity re-targeting (DART) format, has shown promise in the treatment of refractory AML, highlighting the importance of the immune microenvironment in AML treatment (9).
Rationale and knowledge gap
CD69, a membrane-bound type II C-lectin receptor, expressed on different leukocytes, plays a significant immunoregulatory role in the TME (10). For instance, CD69 on tumor-infiltrating cells has been found to be associated with neuroblastoma suppression by simultaneous programmed death 1 (PD-1) and programmed death ligand 1 (PD-L1) blockade (11). The intrahepatic CD69+ Vδ1 T cells could re-circulate in the peripheral blood and limited tumor progression (12). Moreover, CD69 has been identified as a direct hypoxia-inducible factor-1α (HIF-1α) target gene in hypoxia, contributing to its upregulation in tumor-infiltrating T lymphocytes (13). In the treatment of B-cell cancers, T-cells engineered with a chimeric antigen receptor (CAR-T) targeting B-cell activating factor (BAFF) have shown effectiveness in eliminating a range of B-cell malignancies. This process activates large amounts of CD69 and other pro-inflammatory cytokines, effectively diminishes the immune escape of tumor cells (14). In multiple myeloma (MM), a balanced regulation of the immune response is maintained by both CD69+ and CD69− terminal effector T (TTE) cells. Notably, CD69+ TTE cells exhibit elevated expression of inhibitory checkpoints such as CD279, T cell immune receptor with immunoglobulin and immunoglobulin and tyrosine-based inhibitory motif domains (TIGIT), lymphocyte activation gene-3 (Lag3), and CD160. This suggests that CD69+ TTE cells may serve as an optimal immune checkpoint inhibitor target in cancer therapy (15). While the majority of the research on CD69 has centered around B-cell cancers, its involvement in AML is not well understood. Antony et al. reported that CD69 marks a subpopulation of AML with enhanced colony forming capacity and a unique signaling activation state (16). However, further investigation is required to elucidate the relationship between CD69 expression and the pathophysiology of AML.
Objective
This study aimed to analyze the heterogeneous gene expression landscape of AML patients using public databases, focusing on the prognostic implications of this heterogeneity. Our analysis identified 13 hub genes with significant expression alterations in AML. Crucially, the expression of ITGB7, SCD, and CD69 was found to substantially affect the OS of AML patients undergoing decitabine (DA) treatment. Furthermore, we observed a correlation between OS impact and the infiltration levels of B cells, CD4+ T cells and endothelial cells. These findings provide valuable insights into the immune microenvironment of AML and could pave the way for novel drug target discoveries in AML treatment. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1550/rc).
Methods
Data sourcing
This study utilized three gene datasets from the Gene Expression Omnibus (GEO) database: GSE92778, GSE84881, and GSE30029. GSE92778 included RNA sequence data from 6 AML patients and 6 healthy individuals, GSE84881 from 19 AML patients and 4 healthy individuals, and GSE30029 from 46 AML patients and 31 healthy individuals. Additional datasets, GSE40442 and GSE40870, containing RNA sequence data from AML patients treated with cytosine arabinoside (Ara-C) and DA, were also sourced from GEO. The study further incorporated RNA sequence data from AML patients in The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and Therapeutically Applicable Research to Generate Effective Treatments (TARGET) (https://ocg.cancer.gov/programs/target) databases. The expression of specific genes in different tumor cell lines was analyzed using the Cancer Cell Line Encyclopedia (CCLE) (https://sites.broadinstitute.org/ccle/) database.
Data handing
Primary RNA sequence data from different databases were normalized using the “limma” package in R software (version 4.0.3). The expression matrix, including probe ID, was substituted by the corresponding gene ID. Genes with |log2 fold change (FC) >1| and P<0.05 were considered differently expressed genes (DEGs). Volcano plots for identifying DEGs were generated using the “ggplot2” package in R software. Additionally, clinical data, such as patient age, gender, and treatment regimen, were also downloaded from GEO, TCGA and TARGET databases.
Identification and validation of hub genes
Hub genes were identified as DEGs with a P value under 0.05, found across GSE92778, GSE84881, and GSE30029, and showing consistent expression trend (either upregulation or downregulation). The “bioinformatics” online web tool (http://www.bioinformatics.com.cn/) was utilized for this identification and a volcano map of hub genes expression was created. Pathway analysis of these hub genes was performed using the online tool Metascape (http://metascape.org/), with results visualized in the bubble map using “ggplot2” package in R software. Additional analyses were conducted using University of Alabama at Birmingham Cancer (UALCAN) (http://ualcan.path.uab.edu/) and Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) online tools.
Cox regression model and survival analysis
The impact of hub genes on AML patient OS was assessed using Cox proportional hazards regression models with TCGA data and the “forestplot” package in R software. Forest map and nomograms were constructed to visualize these results, accompanied by a calibration curve to verify nomogram accuracy. Furthermore, the influence of hub genes on patient OS was analyzed using the GEPIA online web tool.
Immune cell infiltration analysis
The correlation between CD69 and immune cell infiltration was determined using Spearman’s rank correlation coefficient, based on TCGA immune infiltration data. The correlation P value, correlation coefficient and correlation calculation method were presented on the top of the figures.
Mutation annotation analysis
Mutation data for each hub gene in AML was downloaded from the TCGA database. Mutation classification, mutation type and the top mutation rate of hub genes in AML was analyzed. Mutation annotation formats were created using the “maftool” package in R software.
Cell culture
The Kasumi-1, HL-60, KG1α cell lines were purchased from the Cell Bank, Chinese Academy of Sciences (Shanghai, China) and were characterized using Short Tandem Repeat (STR) markers. Kasumi-1 cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Massachusetts, USA). HL-60 and KG1α cells were cultured in IMDM (Gibco; Thermo Fisher Scientific, Inc.). All media were supplemented with 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and 10% or 20% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). All cell lines were cultured in a humidified atmosphere of 5% CO2 at 37 ℃ and were found to be negative for mycoplasma contamination.
Western blot analysis
The identification of AML cell lines expressing CD69 was accomplished by navigating through the CCLE database (https://portals.broadinstitute.org/ccle). Western blot analysis was employed to validate the protein expression levels of CD69 in these selected cell lines.
For the Western blot procedure, total protein extraction was performed using radio immunoprecipitation assay (RIPA) buffer (Epizyme Biotech, Shanghai, China, cat. No. P1101) containing protease inhibitor cocktail (Beyotime Biotechnology, Shanghai, China, cat. No. P1005) The bicinchoninic acid (BCA) assay method (Beyotime Biotechnology, cat. No. P0011) was employed to detect the protein concentration. Equal amounts of total protein (30 µg) were then separated by protein electrophoresis on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were probed with the indicated antibodies at a 1:1,000 dilution. Protein bands were detected and quantified by Amersham Imager 600 (GE Healthcare, California, USA).
Patients and samples
A total of 29 bone marrow samples (14 AML patients and 15 normal controls) were recruited from Tongji Hospital of Tongji University. The diagnosis of AML was defined according to World Health Organization (WHO) criteria (17). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Medical Ethic Committee of Tongji Hospital of Tongji University on March 2022 (No. K-2022-004). Written informed consent was obtained from each participant.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Mononuclear cells are isolated from bone marrow using density gradient separation (Percoll, Solarbio Life Sciences, Beijing, China). Total RNA was extracted from bone marrow mononuclear cells using Trizol reagent (Thermo Fisher Scientific, Inc.) following the manufacturer’s instruction. Total RNA from each sample was quantified by the NanoDrop ND-1000 and RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Complementary deoxyribonucleic acid (cDNA) was produced by Evo M-MLV RT Kit for qRT-PCR (Accurate Biotechnology, Hunan, China). The Evo M-MLV RT-PCR Kit (Accurate Biotechnology) was used to detect relative messenger RNA (mRNA) expression by an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Massachusetts, USA) with 40 cycles of PCR thermocycling. The primer sequences were as follows: CD69 forward, 5’-ATTGTCCAGGCCAATACACATT-3’ and reverse, 5’-CCTCTCTACCTGCGTATCGTTTT-3’; GAPDH forward, 5’-GGAGCGAGATCCCTCCAAAAT-3’ and reverse, 5’-GGCTGTTGTCATACTTCTCATGG-3’. The mRNA expression of the target gene was calculated relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression.
Statistical analysis
Data analysis was performed using R version 4.0.3 (Institute for Statistics and Mathematics, Vienna, Austria; https://www.r-project.org). The Wilcoxon test was used for two-group comparisons, and the Kruskal-Wallis test for multi-group comparisons. Statistical analyses utilized hazard ratios (HRs), 95% confidence intervals (CIs), and P values as metrics. Significance was set at a two-tailed P value <0.05.
Results
Identification of 13 hub genes with aberrant expression in AML patients compared to healthy individuals
Through analysis of three GEO databases (GSE92778, GSE84881, and GSE30029) from AML patients and healthy individuals, we identified 13 hub genes with aberrant expression. The specific expression of the 13 hub genes in the three datasets were reflected in Table S1. Volcano maps and Venn diagrams revealed eight upregulated and five downregulated genes (Figure 1A-1E). The bubble plots revealed that these hub genes were predominantly involved in the pathways related to protein hydrolysis regulation, cell cycle phase transition, and cell division regulation (Figure 1F).

Identification of 10 hub genes with significant different expression level in AML patients from the aforementioned 13 hub genes by using GEPIA online tool
To discern hub genes exhibiting aberrant expression potentially linked to the progression of AML, we conducted a comprehensive analysis of the expression profiles of the 13 hub genes. This investigation was carried out utilizing the GEPIA online tool, enabling a comprehensive exploration of their expression patterns and potential associations with the development of AML. We compared these genes in TCGA and Genotype-Tissue Expression (GTEx) databases which included the RNA-sequence data of AML patients and healthy people, selecting 173 AML patients and 70 normal controls for comparison.
Our findings revealed that, with the exception of TXNIP, PMAIP1, and TFDP1, the remaining 10 genes (ANKRD36B, ITGB7, S100A10, ADD3, CD69, ERAP2, PLK1, TSPAN13, SCD, and AURKA) exhibited noteworthy variations in expression levels between AML patients and the general population (Figure 2). In particular, ANKRD36B, ITGB7, S100A10, ADD3, CD69, and ERAP2 displayed significantly elevated expression in AML patients compared to the general population (P<0.05). Conversely, PLK1, TSPAN13, SCD, and AURKA demonstrated significantly reduced expression in AML patients (P<0.05).

Recognizing that alterations in gene expression are not the sole contributors to AML, we conducted an examination of the mutation status of the 10 significant differently expressed hub genes based on the TCGA database. This dual perspective provides a more comprehensive understanding of the molecular landscape and potential drivers involved in AML. Our comprehensive analysis revealed that these genes predominantly underwent missense mutations upon the onset of AML (Figure S1A). Additionally, our investigation identified single nucleotide polymorphisms (SNPs) within these hub genes (Figure S1B). Our observations highlighted that the most prevalent single nucleotide variant (SNV) in AML entailed the substitution of C with T across these hub genes (Figure S1C). Finally, our findings underscored that the top two genes most frequently subjected to mutations in AML were CD69 and ADD3 (Figure S1D).
Expression of hub genes in patients with different clinical features
We utilized the TCGA database to investigate the association between the expression patterns of the 10 genes and diverse clinical features. The patient cohort was stratified based on age, distinguishing those over 60 years old from the younger counterparts, further subgroups were established based on gender. Our findings illuminated a decrease in the expression levels of ANKRD36B and TSPAN13, coupled with an increase in S100A10, CD69, and SCD expression within the elderly group (Figure 3). Next, we employed the UALCAN online web tool to analyze the expression of these hub genes in patients with different gender. CD69 (P=0.02), PLK1 (P=0.02), SCD (P=0.03) and AURKA (P=0.02) exhibited lower expression levels in the female patients group compared to the male patients group (Figure 4). These results suggest that CD69 and SCD exhibit variable expression levels among patients with different clinical features such as age and gender. To further refine our analysis of these two genes, we downloaded RNA-sequence data from the TARGET database. Although no significant differences in CD69 expression were observed between patients of different genders, CD69 expression was significantly lower in Caucasian patients (Figure S2).


Impact of CD69 on AML patient survival
To investigate the potential impact of the 10 hub genes on the survival outcomes of AML patients, we divided the AML patients into two groups based on the median expression of the genes of interest. This division facilitated a comparative analysis of the expression levels of these genes by employing the GEPIA web tool. The results revealed that the CD69low group exhibited a more favorable OS compared to the CD69high group (Figure 5). Interestingly, the remaining genes did not show a statistically significant association with survival outcomes.

Cox regression model for risk factors in AML
To further explore prognostic biomarkers of AML, we constructed a Cox regression model. In the univariate Cox regression analysis, the expression of ITGB7 (univariate, HR =1.314; 95% CI: 1.109–1.557; P=0.002), SCD (univariate, HR =1.229; 95% CI: 1.068–1.416; P=0.004) and age (univariate, HR =1.042; 95% CI: 1.026–1.058; P<0.001) were identified as risk factors (Figure 6A).

In the multivariate Cox regression analysis, the expression of CD69 (multivariate, HR =1.217; 95% CI: 1.001–1.479; P=0.049), ITGB7 (multivariate, HR =1.217; 95% CI: 1.001–1.479; P=0.049), SCD (multivariate, HR =1.366; 95% CI: 1.131–1.650; P=0.001) and age (multivariate, HR =1.042; 95% CI: 1.024–1.061; P<0.001) were identified as independent risk factors (Figure 6B).
To facilitate a comprehensive understanding of these findings, we designed a nomogram. Higher points for each variable in the nomogram indicate a greater impact on patients’ OS. Our observations underscore that CD69, ITGB7, SCD, and age are pivotal risk factors influencing OS. As the expression of CD69, ITGB7, and SCD increases, the total points increase, correlating with a decreased survival rate at 1, 2, and 3 years, especially for ITGB7 (Figure 6C). Moreover, age is also emerged as a significant risk factor influencing patients’ survival status (Figure 6C). This suggests that the expression of CD69, ITGB7, SCD, and age collectively impacts the OS of AML patients.
Additionally, we constructed a calibration curve (Figure 6D) to assess the nomogram’s predictive accuracy. The calibration curve demonstrated a close alignment between the predicted 1-, 2-, and 3-year survival rates from the nomogram and the actual observed survival rates.
Correlation of CD69 with immune cells infiltration in AML
CD4+ T cell subsets have been implicated in immune regulatory mechanisms that can suppress anti-tumor responses (18). Additionally, CD8+ T cells and macrophages also play crucial roles in anti-tumor immunity and are associated with the prognosis of AML. These findings suggest that CD69 may exert influence on the activity of AML cells through its impact on immune cells infiltration. Therefore, we conducted an analysis of the correlation between CD69 expression and immune cell infiltration in AML using TCGA data.
Our analysis revealed that CD69 expression was positively correlated with the infiltration levels of B cells (P=0.01), CD4+ T cells (P<0.001) and endothelial cells (P=0.001). On the other hand, CD69 expression was negatively correlated with the infiltration levels of CD8+ T cells (P=0.03) and macrophages (P=0.03) (Figure 7).

Impact of CD69 on DA treatment efficacy in AML
The results of the aforementioned studies compellingly demonstrated that CD69 has a significant impact on the occurrence, prognosis, and infiltration of immune cells in AML. Building on these findings, we hypothesized that CD69 might play a crucial role in influencing the efficacy of chemotherapy treatment in AML. To explore this further, we analyzed data from GSE40442 and GSE40870, which provided information on AML patients treated with Ara-C and DA.
In GSE40442, the expression of CD69 decreased in AML patients treated with DA but not in those treated with Ara-C (Figure 8A,8B). Similar trends were observed in GSE40870, indicating that DA could specifically affect the expression level of CD69 (Figure 8C,8D). DA is a commonly used hypomethylating agents in the treatment of AML. Given the typically low remission rates achieved with monotherapy, it was often combined with other drugs for AML treatment.

Validation of CD69 expression in AML cell lines and AML patients
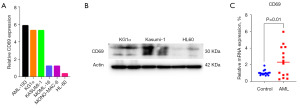
To investigate the expression and function of CD69 in AML cells, we analyzed the cell line expression data from the CCLE database, and our analysis revealed that AML-193 cells exhibited the highest mRNA expression of CD69, followed by KG1α (Figure 9A). These findings were further confirmed by Western blot analysis results (Figure 9B). Further, to validate the expression of CD69 in AML patients, bone marrow mononuclear cells were isolated from AML patients and the normal controls. The expression of CD69 in bone marrow mononuclear cells were detected by qRT-PCR, and the results showed that CD69 was more transcribed in AML patients compared with the normal controls (Figure 9C), which was consistent with the result observed in the TCGA and GTEx databases.
Discussion
Key findings
In this study, 13 hub genes with aberrant expression were identified in AML patients compared to healthy controls. Among these, ANKRD36B, ITGB7, S100A10, ADD3, CD69 and ERAP2 showed significantly increased expression, while PLK1, TSPAN13, SCD and AURKA were significantly downregulated. Our analysis presented a notable decrease in the expression of ANKRD36B and TSPAN13 alongside an increase in S100A10, CD69, and SCD in patients over 60 years old. We highlighted intriguing differences in gene expression between male and female patients. Female patients exhibited lower expression levels of CD69, PLK1, SCD, and AURKA compared to male patients. We also identified that CD69, ITGB7, SCD and age were independent risk factors influencing AML patients’ prognosis, and indicated a strong relationship between CD69 expression and immune cell infiltration particularly an inverse association with CD8+ T cells and macrophages, key players in anti-tumor immunity. Additionally, our study found that DA treatment, a known DNA methylation inhibitor, significantly decreased CD69 expression in AML patients treated with DA.
Comparison with similar researches and explanations of findings
PLK1 and AURKA are important genes in the treatment of AML. The drug volasertib (Vol), a PLK1 inhibitor, has been combined with cytarabine, a classic AML drug, showing promising results (19). Additionally, using a nano-delivery system guided by transferrin to deliver PLK1 inhibitors has greatly improved its effectiveness and safety in AML treatment (20). The AURKA gene translates a crucial kinase responsible for tumor necrosis factor receptor-associated factor (TRAF)-interacting protein with fork head-associated (FHA) domain (TIFA) phosphorylation, leading to the activation of the nuclear factor kappa-B (NF-κB) pathway in AML, promoting disease progression (21). Proteolysis targeting chimeras (PROTACs) against AURKA have been developed to target AML proteins, overcome the “hook effect” and prevent resistance to treatment caused by cancer stem cells (22). Li et al. reported that TET2 deficiency in AML blast cells increases expression of TSPAN13, leading to increased homing/migration of leukemia stem cells into bone marrow niche (23). Long et al. reported that upregulating SCD protects AML cells from ferroptosis and promotes the proliferation (3). However, the role of other hub genes, such as ANKRD36B, S100A10, ADD3, and ERAP2, are not well understood. Further investigation is needed to explore their involvement in the development and progression of AML.
The ITGB family, known for its role as an adhesion receptor, is crucial in organizing the cytoskeleton and influencing various signaling pathways that control cell proliferation (24,25). Our study highlighted the ITGB7 gene, a member of ITGB family, as a significant marker for AML prognosis. Its higher expression in older AML patients and identification as an independent risk factor suggested its utility in predicting AML outcomes. According to previous research, AML cells express multiple integrins including ITGB3, another member of ITGB family. Research has indicated that the Syk kinase plays a key role in regulation the proliferation of AML cell through its regulation of ITGB3. Additionally, overexpression of ITGB3 reduces the sensitivity of the kinase inhibitor sorafenib, resulting in a poor prognosis for AML (26). Our research, as well as the literature, reinforced the importance of the ITGB family in AML.
This study found that CD69 was highly expressed in AML patients, particularly in male and older subgroups. Patients with lower expression of CD69 had longer OS time, while those with increased expression of CD69 was associated with poor prognosis, suggesting that CD69 might serve as a disease prognosticator. Analysis of TCGA datasets also revealed a close relationship between CD69 and immune cell infiltration. Our study showed that CD69 expression was positively correlated with the infiltration levels of B cells, CD4+ T cells, and endothelial cells, while negatively correlated with CD8+ T cells and macrophages. The TME consists of various cells and tissues with different functions, and immune infiltration plays a crucial role in disease progression. For instance, CD8+ T cells mediate cytotoxic responses to kill tumor cells, while macrophages can exert dual influences by either blocking or promoting tumorigenesis. An increase in tumor-associated macrophages is generally associated with poor prognosis and drug resistance (27,28). In our study, CD69 expression was inversely associated with immune cells that inhibit tumor progression under specific conditions. This suggested that AML cells with overexpressed CD69 modulated immune cell infiltration and reduced the expression of CD8+ T cells, macrophages, or other antitumor cells, thereby promoting disease progression and immune escape. Therefore, CD69 is a promising target, and combination therapy targeting CD69 may be a potential new direction for AML treatment.
DA and Ara-C are commonly used in the treatment of AML (29). DA is an inhibitor of DNA methylation which can reactivate tumor suppressor genes (30). Ara-C inhibits DNA synthesis by targeting replicative polymerase epsilon (31). Our present study based on two datasets found that DA influenced CD69 expression while Ara-C did not. The expression level of CD69 receiving DA treatment was lower than that of the group receiving Ara-C therapy. Encouraging results have been observed in children with relapsed/refractory acute leukemia treated with azacitidine and cytarabine (32) and in adults with AML treated with DA and vorinostat (33,34). The differential effects of DA and Ara-C on the expression of CD69 might shed light on further investigations for identifying better combination of epigenetic agents of AML treatments.
Strengths and limitations
Our study showed that CD69 emerged as a prominent factor with its high expression correlating with poor prognosis in AML, provided valuable insights into the immune microenvironment of AML that have the potential to aid in the discovery of drug targets for AML patients. These discoveries underscored the complexity of AML and the need for strategies that considered individual genetic profiles and demographic factors. These insights could pave the way for developing more effective and personalized treatment modalities for AML patients.
This study has some limitations. Firstly, although we initially identified correlations between CD69 and SCD molecules and specific clinical features of AML through analysis of the TCGA database, these associations did not consistently match when validated with RNA sequences from the target database. This discrepancy could be linked to inherent heterogeneity across diverse datasets. Therefore, further validation using larger-scale clinical data is necessary to affirm and clarify these observed differences. Secondly, our study focused on affirming the correlation between CD69 and the immune response in AML. Unfortunately, constraints within the available datasets precluded the selection of suitable normal controls for comparative analysis. Nonetheless, we successfully established a linear correlation between the extent of immune cell infiltration and the magnitude of CD69 molecular alterations. Therefore, further research is imperative to delve into the role of CD69 in the pathogenesis and progression of AML, unraveling its specific mechanisms. This will be the focus of our future research.
Conclusions
Our findings revealed that the expression of ITGB7, SCD, and CD69 were significantly correlated with the OS of AML patients. Additionally, SCD and ITGB7 expression, along with advanced age, were identified as factors influencing poor prognosis in AML. Furthermore, the study established CD69 as a critical biomarker and molecular target in AML progression and immune cell infiltration. Based on our results, CD69 merits further research for a deeper understanding of its role in AML progression and immune regulation.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1550/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1550/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1550/prf
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1550/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethic Committee of Tongji Hospital of Tongji University on March 2022 (No. K-2022-004). Written informed consent was obtained from each participant.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kantarjian H, Kadia T, DiNardo C, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J 2021;11:41. [Crossref] [PubMed]
- Shimony S, Stahl M, Stone RM. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol 2023;98:502-26. [Crossref] [PubMed]
- Long F, Lin Z, Long Q, et al. CircZBTB46 Protects Acute Myeloid Leukemia Cells from Ferroptotic Cell Death by Upregulating SCD. Cancers (Basel) 2023;15:459. [Crossref] [PubMed]
- Kim HS, Kim D, Kim J, et al. SCD and MTHFD2 inhibitors for high-risk acute myeloid leukaemia patients, as suggested by ELN2017-pathway association. Clin Transl Med 2023;13:e1311. [Crossref] [PubMed]
- Laouedj M, Tardif MR, Gil L, et al. S100A9 induces differentiation of acute myeloid leukemia cells through TLR4. Blood 2017;129:1980-90. [Crossref] [PubMed]
- Böttcher M, Panagiotidis K, Bruns H, et al. Bone marrow stroma cells promote induction of a chemoresistant and prognostic unfavorable S100A8/A9high AML cell subset. Blood Adv 2022;6:5685-97. [Crossref] [PubMed]
- Zhong F, Yao F, Cheng Y, et al. m6A-related lncRNAs predict prognosis and indicate immune microenvironment in acute myeloid leukemia. Sci Rep 2022;12:1759. [Crossref] [PubMed]
- Rutella S, Vadakekolathu J, Mazziotta F, et al. Immune dysfunction signatures predict outcomes and define checkpoint blockade-unresponsive microenvironments in acute myeloid leukemia. J Clin Invest 2022;132:e159579. [Crossref] [PubMed]
- Uy GL, Aldoss I, Foster MC, et al. Flotetuzumab as salvage immunotherapy for refractory acute myeloid leukemia. Blood 2021;137:751-62. [Crossref] [PubMed]
- Lin R, Zhang H, Yuan Y, et al. Fatty Acid Oxidation Controls CD8(+) Tissue-Resident Memory T-cell Survival in Gastric Adenocarcinoma. Cancer Immunol Res 2020;8:479-92. [Crossref] [PubMed]
- Inoue S, Takeuchi Y, Horiuchi Y, et al. CD69 on Tumor-Infiltrating Cells Correlates With Neuroblastoma Suppression by Simultaneous PD-1 and PD-L1 Blockade. J Surg Res 2023;289:190-201. [Crossref] [PubMed]
- Bruni E, Cimino MM, Donadon M, et al. Intrahepatic CD69(+)Vδ1 T cells re-circulate in the blood of patients with metastatic colorectal cancer and limit tumor progression. J Immunother Cancer 2022;10:e004579. [Crossref] [PubMed]
- Labiano S, Meléndez-Rodríguez F, Palazón A, et al. CD69 is a direct HIF-1α target gene in hypoxia as a mechanism enhancing expression on tumor-infiltrating T lymphocytes. Oncoimmunology 2017;6:e1283468. [Crossref] [PubMed]
- Wong DP, Roy NK, Zhang K, et al. A BAFF ligand-based CAR-T cell targeting three receptors and multiple B cell cancers. Nat Commun 2022;13:217. [Crossref] [PubMed]
- Vuckovic S, Bryant CE, Lau KHA, et al. Inverse relationship between oligoclonal expanded CD69- TTE and CD69+ TTE cells in bone marrow of multiple myeloma patients. Blood Adv 2020;4:4593-604. [Crossref] [PubMed]
- Antony ML, Chang D, Noble-Orcutt KE, et al. CD69 marks a subpopulation of acute myeloid leukemia with enhanced colony forming capacity and a unique signaling activation state. Leuk Lymphoma 2023;64:1262-74. [Crossref] [PubMed]
- Arber DA, Orazi A, Hasserjian RP, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood 2022;140:1200-28. [Crossref] [PubMed]
- Gournay V, Vallet N, Peux V, et al. Immune landscape after allo-HSCT: TIGIT- and CD161-expressing CD4 T cells are associated with subsequent leukemia relapse. Blood 2022;140:1305-21. [Crossref] [PubMed]
- Zeidan AM, Ridinger M, Lin TL, et al. A Phase Ib Study of Onvansertib, a Novel Oral PLK1 Inhibitor, in Combination Therapy for Patients with Relapsed or Refractory Acute Myeloid Leukemia. Clin Cancer Res 2020;26:6132-40. [Crossref] [PubMed]
- Xia Y, An J, Li J, et al. Transferrin-guided intelligent nanovesicles augment the targetability and potency of clinical PLK1 inhibitor to acute myeloid leukemia. Bioact Mater 2023;21:499-510. [Crossref] [PubMed]
- Wei TW, Wu PY, Wu TJ, et al. Aurora A and NF-κB Survival Pathway Drive Chemoresistance in Acute Myeloid Leukemia via the TRAF-Interacting Protein TIFA. Cancer Res 2017;77:494-508. [Crossref] [PubMed]
- Liu F, Wang X, Duan J, et al. A Temporal PROTAC Cocktail-Mediated Sequential Degradation of AURKA Abrogates Acute Myeloid Leukemia Stem Cells. Adv Sci (Weinh) 2022;9:e2104823. [Crossref] [PubMed]
- Li Y, Xue M, Deng X, et al. TET2-mediated mRNA demethylation regulates leukemia stem cell homing and self-renewal. Cell Stem Cell 2023;30:1072-1090.e10. [Crossref] [PubMed]
- Pang X, He X, Qiu Z, et al. Targeting integrin pathways: mechanisms and advances in therapy. Signal Transduct Target Ther 2023;8:1. [Crossref] [PubMed]
- Kamranvar SA, Rani B, Johansson S. Cell Cycle Regulation by Integrin-Mediated Adhesion. Cells 2022;11:2521. [Crossref] [PubMed]
- Johansen S, Brenner AK, Bartaula-Brevik S, et al. The Possible Importance of β3 Integrins for Leukemogenesis and Chemoresistance in Acute Myeloid Leukemia. Int J Mol Sci 2018;19:251. [Crossref] [PubMed]
- Bejarano L, Jordāo MJC, Joyce JA. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov 2021;11:933-59. [Crossref] [PubMed]
- DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369-82. [Crossref] [PubMed]
- Ling VY, Straube J, Godfrey W, et al. Targeting cell cycle and apoptosis to overcome chemotherapy resistance in acute myeloid leukemia. Leukemia 2023;37:143-53. [Crossref] [PubMed]
- Penter L, Liu Y, Wolff JO, et al. Mechanisms of response and resistance to combined decitabine and ipilimumab for advanced myeloid disease. Blood 2023;141:1817-30. [Crossref] [PubMed]
- Owen N, Minko IG, Moellmer SA, et al. Enhanced cytarabine-induced killing in OGG1-deficient acute myeloid leukemia cells. Proc Natl Acad Sci U S A 2021;118:e2016833118. [Crossref] [PubMed]
- Stahl M, Menghrajani K, Derkach A, et al. Clinical and molecular predictors of response and survival following venetoclax therapy in relapsed/refractory AML. Blood Adv 2021;5:1552-64. [Crossref] [PubMed]
- How J, Minden MD, Brian L, et al. A phase I trial of two sequence-specific schedules of decitabine and vorinostat in patients with acute myeloid leukemia. Leuk Lymphoma 2015;56:2793-802. [Crossref] [PubMed]
- Kirschbaum M, Gojo I, Goldberg SL, et al. A phase 1 clinical trial of vorinostat in combination with decitabine in patients with acute myeloid leukaemia or myelodysplastic syndrome. Br J Haematol 2014;167:185-93. [Crossref] [PubMed]


